
Gene expression trajectories aiding the unveiling of phenotypic similarities in developmental disorders: insights from human-mouse comparative analysis
Highlight box
Key findings
• In genes associated with developmental disorders, viable with phenotype (VP) genes with similar expression trajectories in the brain exhibited analogous phenotypes across human and mouse.
What is known and what is new?
• Knockout mouse models are critical tools in diseases research, but null mutations of orthologous genes in human and mouse often resulted in different phenotypes.
• The gene expression trajectory offered a potential elucidation for the disparate phenotypes observed due to null mutations in humans and mice.
What is the implication, and what should change now?
• Currently, we have gained insights into the ability of gene expression trajectories to unveil the underlying causes of diverse phenotypes resulting from mutations in humans and mice. Future studies are needed to explore the practical application of gene expression trajectories in predicting candidate pathogenic genes.
Introduction
Major congenital malformations and/or severe neurodevelopmental disorders are prevalent in approximately 2–5% of the pediatric population (1,2). These conditions are diseases of impaired prenatal development and are primarily caused by deleterious genetic variations in genes that are crucial for proper development (3). Understanding the genetic basis of these disorders is crucial for developing effective treatments and interventions. Knockout mouse models have emerged as valuable tools in the biomedical sciences for investigating gene-disease associations (4), assuming that orthologous genes a in mice and humans share similar functions. However, it is worth noting that a considerable number of genes exhibit different phenotypes between humans and mice. Previous studies using knockout mouse models have shown that a considerable percentage (49.53%) of genes linked to disorders do not exhibit a shared phenotype between the mouse model and the corresponding human disease (4). This discrepancy may be attributed to functional divergence during evolution.
Genes associated with the developmental process are precisely regulated (5). Species with comparable developmental trajectories often display shared patterns in conserved regulatory networks and molecular processes that contribute to observed phenotypic similarities. Previous studies have revealed that half of human genes showed distinct temporal trajectories comparing to their mouse orthologs (6). Zheng-Bradley et al., found no significant contribution of expression level divergence between human and mouse orthologous genes across homologous tissues of the two species from microarray expression datasets (7). It’s unclear whether the similarity of developmental expression trajectories could serve as an informative index for phenotypic similarity as widely assumed.
Developmental expression trajectories refer to the dynamic patterns of gene expression throughout the process of development. Similarity of expression trajectories can be quantified by comparing RNA sequencing (RNA-seq) data from homologous tissues of human and mouse across calibrated developmental ages (8). However, quantifying phenotype similarities between species poses a significant challenge. The Online Mendelian Inheritance in Man (OMIM) project employs the Human Phenotype Ontology (HPO) to describe disease phenotypes in a structured hierarchy of terms (9). The International Mouse Phenotyping Consortium (IMPC) generates a vast number of mouse models that are extensively screened for detailed phenotypes (10). These phenotype data are captured and organized using the Mammalian Phenotype Ontology (MPO) (11). By leveraging semantic similarities (12) in quantifying the similarities between HPO and MPO, we can bridge the gap in associations between similarities in expression trajectories and similarities in phenotypes.
In this study, we explored whether genes associate with developmental disorders with similar expression trajectories exhibit similar phenotypes, and to determine whether this pattern is general or specific to a particular set of genes. To accomplish this, we probed the relationship between expression trajectory similarities and phenotypic similarities between humans and mice using genes from Developmental Disorders Genotype-to-Phenotype (DDG2P) database (3). Additionally, our study is centered on the gene family of ionotropic glutamate receptors (iGluRs), which has been linked to neurodevelopmental disorders. Specifically, we investigated the impact of protein-truncating variants (PTVs) in both human and mouse models to exclude the potential confounding effects of distinct functions, and further elucidate the feasibility of inferring similar phenotypes based on similar expression trajectories. Our study aims to provide an expanded perspective on comprehending developmental diseases and the discovering the potential disease-related genes.
Methods
Research procedure
In order to investigate the feasibility of utilizing mouse models to simulate human developmental disorders, we employed the HPO and MPO terms to compute original Phenodigm scores, serving as indicators of phenotypic similarity. Furthermore, we conducted Spearman correlation coefficient analysis on temporal gene expression data from corresponding tissues in humans and mice to assess the similarities in gene expression trajectories. To mitigate potential confounding effects from variants with diverse functionalities, we meticulously screened cases involving PTVs in the iGluRs gene family, utilizing the HPO terms. Furthermore, we employed the MPO terms in gene knockout mouse models. Through the computation of refined Phenodigm scores based on HPO terms and MPO terms, our study aimed to elucidate the feasibility of harnessing gene expression trajectory similarity as a valuable tool to facilitate the prediction of phenotypic similarities.
Transcriptomic data of human and mouse organ development
We utilized gene temporal expression data from bulk transcriptomes of humans and mice across developmental stages (8). Human data covered lifespan from 8 weeks post-conception (WPC) to 41 years of age. Mouse data covered lifespan from E10.5 to postnatal day (P) 63 and were binned into 12 time points corresponding to human developmental stages (4, 5, 7, 8, 11, 12, 13, 19, 20 WPC and toddler, young adult and young middle age) respectively. 17,150 one-to-one humans to mouse orthologous gene pairs were extracted from the Ensembl (version 107) database (https://www.ensembl.org). Expression similarities were calculated as Spearman correlation ρ (13) between expressions values (measured as reads per kilobase per millions, RPKMs) of human and mouse orthologous genes in the same tissues across the 12 time points. ρ is defined as:
where di is the difference in paired ranks of expression values between human and mouse and n is the number of time points. ρ is ranges from −1 to 1 with the values closer to 1 indicating a stronger positive association between human and mouse RPKMs.
To visualize the gene expression trends in human and mouse tissues, we used relative expression value normalized by the maximum values in tissues. Data were processed using R package tidy verse (14). Plots were generated by the R package ggplot2 (15).
Literature search
We conducted a literature search on PubMed (www.ncbi.nlm.nih.gov/pubmed) using the search terms “GRIA1, GRIA2, GRIA3, GRIA4, GRIK1, GRIK2, GRIK3, GRIK4, GRIK5, GRIN1, GRIN2A, GRIN2B, GRIN2C, GRIN2D, GRIN3A, GRIN3B, GRID1 and GRID2” in July 2022. We reviewed the clinical and genetic information of 213 cases with PTVs in GRIA1, GRIA2, GRIA3, GRIA4, GRIK2, GRIN1, GRIN2A, GRIN2B, GRIN2D and GRID2. PTVs are defined as variants [stop-gained single-nucleotide variants (SNVs), frameshift insertions or deletions, large structural variants and splice-region SNVs] that disrupt transcription and contribute to shortened or absent protein (16). Patients with gross deletion/translocation/insertion variants exceeding the gene length were excluded.
Human and mouse phenotype comparison
The human clinical phenotypes encoded as HPO terms associated with human genes, were downloaded from the HPO website (17) (www.human-phenotype-ontology.org). The HPO terms were initially created by developing an ontology using information from the Clinical Synopsis of the OMIM database and gradually collaborating with clinicians in workshops to enhance and expand the clinical terminology (9). Phenotypes of mouse for each gene were described using MPO terms obtained from MGI database (18) (www.informatics.jax.org/). The MPO serves as a structured vocabulary, allowing the consistent annotation of mouse phenotypic data from various sources (e.g., published literature, large-scale mutagenesis centers, individual research laboratories) using standardized phenotype terminology (19). Original Phenodigm scores (12) generated by HPO terms and MPO terms for human and mouse gene pairs were extracted from BioLink API (v1.1.14) (https://api.monarchinitiative.org/api).
The phenotype similarities for genes with PTVs in iGluRs genes were calculated as refined Phenodigm scores for manually curated HPO and MPO profiles which were stratified on the genotype (homozygous versus heterozygous) of reported cases and the genotype of mouse models (available online: https://cdn.amegroups.cn/static/public/pm-23-27-1.xlsx, https://cdn.amegroups.cn/static/public/pm-23-27-2.xlsx). For patients harboring PTVs, we extracted clinical phenotypes mentioned in the identified articles and cross-referenced them with the HPO database to assign appropriate HPO terms. Two genetic counsellors specialized in neurogenetics performed the phenotype-to-HPO conversion independently and a senior genetic counsellor reviewed and resolved any discrepancies through consensus discussions. The official HPO website was utilized to obtain relevant HPO terms associated with the specific gene under investigation. Subsequently, HPO terms that closely matched the clinical descriptions documented in the literature were meticulously selected for analysis. We also curated the MPO terms for gene knockout mice with documented literature reports in the MGI database and verified them within the manuscript. We used the original Phenodigm scores of all variants as a reference for iGluRs genes without PTV.
Disease gene list and gene classification
Genes associated with developmental disorders were downloaded from the DDG2P database (https://www.ebi.ac.uk/gene2phenotype/downloads). The database was created by Deciphering Developmental Disorders (DDD) Study, which has recruited >13,400 individuals with undiagnosed severe and/or extreme developmental disorders from the UK and Ireland. The main objective of the project is to define the genetic architecture of developmental disorders, while also aiming to identify new developmental disorders loci (20). A total of 2,187 genes, which have a one-to-one mouse ortholog, were used for expression and phenotype comparison. Genes were classified according to full spectrum of intolerance to loss-of-function (FUSIL) bin (21), which took advantage of the comprehensive organismal viability screen performed by the IMPC database (https://www.mousephenotype.org/understand/data-collections/essential-genes-portal/) and the cellular viability studies conducted by Project Achilles Avana data set (https://depmap.org/portal/achilles/) (available online: https://cdn.amegroups.cn/static/public/pm-23-27-3.xlsx).
Statistical analysis
Gene expression trajectory similarities were calculated using Spearman correlation using the ‘cor’ function in R (version 4.2.2) and phenotypic similarities were generated by Phenodigm scores. Wilcoxon test was employed to compare gene expression trajectory similarities or phenotypic similarities between group by using the R package “rstatix” (22) and its “pairwise_wilcox_test” function. The alternative hypothesis was two-sided. To control for multiple comparisons, the Bonferroni adjustment was applied to maintain the overall significance level. Fisher’s exact test were conducted to explore the correlation between DDG2P genes and genes that have similar expression patterns in a specific tissue by using the “fisher.test” function in R package “stats” (23). Chi-squared test was used to compare proportions among groups by using the “chisq_test” function in R package “rstatix” (22). P values of <0.05 were considered statistically significant.
Results
Similarity of gene expression trajectory across human and mouse tissues
In this study, we examined the expression data for the orthologous genes between the human and mouse genomes. Correlation analysis revealed the orthologous genes were more similarly expressed in the brain and cerebellum than in the other tissues (Figure 1A). Given that gene expression undergoes dynamic changes during the developmental process, we conducted an analysis of the gene expression trajectories similarities between orthologous DDG2P genes and non-DDG2P genes in various tissues to investigate the genes related to developmental disorders. In most major organs including the brain (Z=−0.04, P<0.001, Wilcoxon test), cerebellum (Z=−0.02, P=0.002, Wilcoxon test), heart (Z=−0.04, P<0.001, Wilcoxon test), kidneys (Z=−0.02, P=0.02, Wilcoxon test), liver (Z=−0.02, P<0.001, Wilcoxon test), and testes (Z=−0.02, P<0.001, Wilcoxon test), the gene expression trajectory similarities of DDG2P genes across humans and mice were higher than that of non-DDG2P genes (Figure 1B). Furthermore, orthologous genes displaying similar expression patterns in the brain (OR =1.31, 95% CI: 1.44–1.20, P<0.001), cerebellum (OR =1.15, 95% CI: 1.04–1.27, P=0.006), heart (OR =1.32, 95% CI: 1.19–1.46, P<0.001), kidneys (OR =1.14, 95% CI: 1.02–1.27, P=0.02), and testes (OR =1.25, 95% CI: 1.12–1.39, P<0.001) between humans and mice were significantly enriched within the DDG2P genes (Figure 1C).
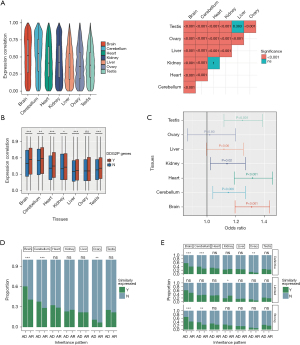
Upon categorizing DDG2P genes into autosomal dominant (AD) genes and autosomal recessive (AR) genes, it was observed that AD genes exhibit a substantially higher proportion of genes displaying similar expression trajectories of the brain (60.2% vs. 40.7%, P<0.001, chi-squared test) and cerebellum (37.6% vs. 28.3%, P<0.001, chi-squared test) in comparison to AR genes (Figure 1D). Furthermore, after stratifying genes according to their degree of association with the disease, AD genes consistently displayed a higher proportion of genes with similar expression trajectories in the brain and cerebellum, specifically within the subset of genes displaying a definitive (brain: 57.3% vs. 38.8%, P<0.001; cerebellum: 38.2% vs. 28.1%, P<0.001, chi-squared test) and strong (brain: 66.8% vs. 42.5%, P<0.001; cerebellum: 38.2% vs. 25.5%, P=0.003, chi-squared test) correlation with developmental delay (Figure 1E).
Phenotypic similarity across human and mouse
The aim of our study is to elucidate the potential impact of similarity in gene expression trajectories on phenotypic similarity. Considering that genes associated with lethal mouse phenotypes are enriched in DDG2P genes (21), we categorized the orthologous DDG2P gene based on full spectrum of FUSIL categorization (21) to mitigate phenotypic dissimilarities arising from genes belonging to different categories (Table 1). This categorization selected human orthologous genes with viability assessments from the IMPC and integrated their essentiality characterization derived from cell proliferation scores obtained from Project Achilles Avana CRISPR-Cas9 screening performed on over 400 cell lines (21). FUSIL categorization classified genes into eight categories based on features observed in both the mouse phenotypes and human cells. After discarding categories with relatively low numbers of genes, categorization was left with a final set of five gene clusters (21).
Table 1
| Mouse category | Human cell line category | Number of orthologous DDG2P genes (n=865) | FUSIL category (20) |
|---|---|---|---|
| Lethal | Essential | 145 | Cellular lethal |
| Lethal | Non-essential | 321 | Developmental lethal |
| Subviable | Essential | – | – |
| Subviable | Non-essential | 158 | Subviable |
| Viable with phenotypic abnormalities | Essential | – | – |
| Viable with phenotypic abnormalities | Non-essential | 225 | Viable with phenotype |
| Viable with normal phenotype | Essential | – | – |
| Viable with normal phenotype | Non-essential | 13 | Viable with no phenotype |
DDG2P, Developmental Disorders Genotype-to-Phenotype; FUSIL, full spectrum of intolerance to loss-of-function.
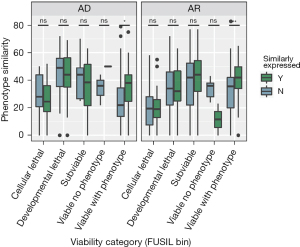
The genes meeting the criteria of being non-essential in human and displaying viable and abnormal phenotypes in knockout mouse models are collectively referred to as “VP genes”. By comparing the phenotypic similarities between humans and mice across the five gene clusters, we found that VP genes exhibiting similar expression trajectories in the brain displayed more similar phenotypes than those with dissimilar expression trajectories (AD genes: Wilcoxon test: Z=11, P=0.02; AR genes: Wilcoxon test: Z=9, P=0.003) (Figure 2).
Similarity of expression trajectory of iGluRs genes in the brain and cerebellum
Genes belonging to the same family have similar sequences and possibly the same evolutionary origins and similar functions (24). The gene expression trajectories of orthologous genes in the brain and cerebellum manifested a pronounced convergence between humans and mice and neurodevelopmental disorders arises from abnormalities in brain development (25). Therefore, using a neurodevelopmental disorder-associated gene family as an example, we explore the feasibility of utilizing gene expression trajectory similarity to infer phenotypic similarity.
The correlation coefficients of expression trajectory similarity between human and mouse were shown in Table 2. Ten of the iGluRs disease-causing genes have been reported in the OMIM database (Table 3), which were also incorporated into the DDG2P database. Within the disease-causing gene set, we observed that GRIA4 (brain: ρ=0.80, P=0.003; cerebellum: ρ=0.79, P=0.03), GRIN1 (brain: ρ=0.93, P<0.001; cerebellum: ρ=0.86, P=0.01), GRIN2A (brain: ρ=0.83, P=0.001; cerebellum: ρ=0.88, P=0.007) and GRIN2B (brain: ρ=0.76, P=0.007; cerebellum: ρ=0.88, P=0.007) displayed comparable expression trajectories in both brain and cerebellum across humans and mice (Figure S1A-S1D). Moreover, GRIA2 (ρ=0.78, P=0.005) and GRIA3 (ρ=0.93, P<0.001) exhibited similar expression trajectories solely within the brain of humans and mice (Figure S1E,S1F). Conversely, the remaining disease-causing genes (GRIA1, GRIK2, GRIN2D and GRID2) exhibited divergent expression trajectories within the brain and cerebellum across species (Figure S1G-S1J).
Table 2
| Receptor | Gene | Inheritance | Number of cases with PTVs (n=213) | Number of cases with missense variants (n=332) | OMIM number |
|---|---|---|---|---|---|
| AMPA receptor | GRIA1 | AD | – | 6 | 619927 |
| AR | 1 | – | 619931 | ||
| GRIA2 | AD | 5 | 21 | 618917 | |
| GRIA3 | XLR | – | 16 | 300699 | |
| GRIA4 | AD | – | 6 | 617864 | |
| KA receptor | GRIK2 | AD | – | 12 | 619580 |
| AR | 14 | – | 611092 | ||
| NMDA receptor | GRIN1 | AD | – | 48 | 614254 |
| AR | 4 | – | 619814 | ||
| AR | 614254 | ||||
| GRIN2A | AD | 146 | 122 | 245570 | |
| GRIN2B | AD | 26 | 82 | 616139 | |
| AD | 613970 | ||||
| GRIN2D | AD | – | 13 | 617162 | |
| Delta receptor | GRID2 | AR | 17 | 6 | 616204 |
iGluRs, ionotropic glutamate receptors; PTVs, protein-truncating variants; OMIM, Online Mendelian Inheritance in Man; AMPA, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid; KA, kainite; NMDA, N-methyl-d-aspartate; AD, autosomal dominant; AR, autosomal recessive; XLR, X-linked recessive.
Table 3
| Relationship between genes and diseases | Gene | Brain | Cerebellum | Similar gene expression trajectories | |||
|---|---|---|---|---|---|---|---|
| Correlation coefficient (Spearman) | P value | Correlation coefficient (Spearman) | P value | ||||
| Disease-causing genes | GRIA4 | 0.80 | 0.003 | 0.79 | 0.03 | Brain and cerebellum | |
| GRIN1 | 0.93 | 0 | 0.86 | 0.01 | Brain and cerebellum | ||
| GRIN2A | 0.83 | 0.001 | 0.88 | 0.007 | Brain and cerebellum | ||
| GRIN2B | 0.76 | 0.007 | 0.88 | 0.007 | Brain and cerebellum | ||
| GRIA2 | 0.78 | 0.005 | 0.12 | 0.79 | Brain | ||
| GRIA3 | 0.93 | 0 | −0.52 | 0.20 | Brain | ||
| GRIA1 | 0.31 | 0.32 | 0.33 | 0.43 | – | ||
| GRIK2 | 0.56 | 0.06 | 0.50 | 0.22 | – | ||
| GRIN2D | 0.47 | 0.13 | 0.31 | 0.46 | – | ||
| GRID2 | 0.2 | 0.43 | 0.62 | 0.12 | – | ||
| Unknown disease-causing genes | GRIK3 | 0.87 | <0.001 | 0.83 | 0.02 | Brain and cerebellum | |
| GRIN2C | 0.91 | 0 | 0.81 | 0.02 | Brain and cerebellum | ||
| GRIK4 | 0.81 | 0.002 | 0.55 | 0.17 | Brain | ||
| GRID1 | 0.93 | 0 | 0.36 | 0.39 | Brain | ||
| GRIK1 | 0.43 | 0.16 | 0.86 | 0.01 | Cerebellum | ||
| GRIN3A | 0.37 | 0.24 | 0.81 | 0.02 | Cerebellum | ||
| GRIK5 | 0.29 | 0.37 | 0.48 | 0.24 | – | ||
| GRIN3B | −0.55 | 0.07 | 0.62 | 0.12 | – | ||
iGluRs, ionotropic glutamate receptors.
Regarding genes with unknown disease-causing potential, our study detected similar expression trajectories in the brain and cerebellum between humans and mice for GRIK3 (brain: ρ=0.87, P<0.001; cerebellum: ρ=0.83, P=0.02) and GRIN2C (brain: ρ=0.91, P<0.001; cerebellum: ρ=0.81, P=0.02) (Figure S2A,S2B). The GRIK4 (ρ=0.81, P=0.002) and GRID1 (ρ=0.93, P<0.001) displayed matching expression trajectories in the brain of humans and mice (Figure S2C,S2D). And the GRIK1 (ρ=0.86, P=0.01) and GRIN3A (ρ=0.81, P=0.02) expressed analogous trajectories in the cerebellum across species (Figure S2E,S2F). However, the expression trajectories of the remaining genes (GRIK5, GRIN3B) differed in the brain and cerebellum between humans and mice (Figure S2G,S2H).
Phenotypic similarity of disease-causing iGluRs genes
Out of the ten disease-causing genes of iGluRs, GRIA1 was classified as a VP bin, GRIA2 as developmental lethal, GRIN2B as subviable, and the remaining seven disease-causing genes were not included in the FUSIL categorization dataset. Homozygous null variants of GRIA2, GRIN1, and GRIN2B were found to result in lethal phenotypes (26-28). The VP category defined the knockout mouse were viable and presented significantly differing phenotypes in at least one of parameters (21), which was applicable to the remaining genes. Currently, none of the genes within a gene family in the DDG2P database have been classified entirely as VP genes.
As is known, variants of iGluRs can lead to alterations in function, including gain of function and loss of function. To eliminate the potential interference caused by functional alterations, we conducted a review of previous studies on cases with PTVs of iGluR genes to calculate refined Phenodigm scores (available online: https://cdn.amegroups.cn/static/public/pm-23-27-2.xlsx). PTVs are defined as variants [stop-gained single-nucleotide variants (SNVs), frameshift insertions or deletions, large structural variants and splice-region SNVs] that disrupt transcription and contribute to a shortened or absent protein, which was considered to have the loss of function effect. PTVs of GRIA3, GRIA4, and GRIN2D had not been previously reported in neurodevelopmental disorders. Therefore, we used the original Phenodigm scores of all variants as a reference in our analysis.
For genes with similar expression trajectories in the brain and cerebellum across humans and mice, the GRIA4 [56] and GRIN1 [67] respectively showed the highest original Phenodigm score and the highest refined Phenodigm score (Table 4). The refined Phenodigm score of homozygous GRIN2A mice models is 47, and the heterozygous GRIN2B [49] mouse model exhibited a higher refined Phenodigm score than the homozygous [39] (Table 4). In summary, the mouse lacking iGluRs genes with similar expression trajectories in both the brain and cerebellum showed similar phenotypes to human DD.
Table 4
| Gene | Original Phenodigm score | Refined Phenodigm score | |
|---|---|---|---|
| Hom model | Het model | ||
| GRIA1 | – | 60 | – |
| GRIA2 | 54 | 27 | 35 |
| GRIA3 | 34 | – | – |
| GRIA4 | 56 | – | – |
| GRIK2 | 47 | 53 | – |
| GRIN1 | 48 | 67 | – |
| GRIN2A | 41 | 47 | – |
| GRIN2B | 50 | 39 | 49 |
| GRIN2D | 27 | – | – |
| GRID2 | 53 | 53 | – |
iGluRs, ionotropic glutamate receptors.
In regard to genes with divergent expression trajectories within the brain and cerebellum across species, our analysis of human and mouse phenotypes has shown that the correlation between animal models and human disease is weak in GRIN2D [27], while it is robust in GRIA1 [60], GRID2 [53], and GRIK2 [53] with high refined Phenodigm scores.
The GRIA2 gene showed comparable expression trajectories exclusively in the brains of humans and mice and received a relatively low refined Phenodigm score of 27 in homozygous models and 35 in heterozygous models. The low similarity of phenotypes is specifically manifested in impaired coordination. In the GRIA2 knockout mouse model, manifestations of compromised motor coordination were observed (26), while in individuals with PTVs, cerebellar imaging and motor function were reportedly normal (29).These findings suggested that differential expression trajectories of GRIA2 in the cerebellum may contribute to the varied phenotypes between humans and mice.
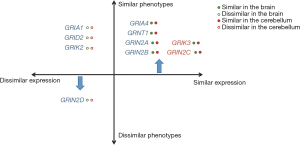
In general, we observed the similar expression trajectories of iGluRs genes in the brain and cerebellum between humans and mice could predict the similar neurological phenotypes across species (Figure 3). Moreover, for the iGluRs genes with dissimilar expression trajectories between the brain and cerebellum, phenotypic similarities depended on whether the phenotypes correlated with organ function.
Discussion
The main objective of this study is to investigate the relationship between the similarities of gene expression trajectory and phenotype in humans and mice for genes associated with DD. Firstly, we compared the gene expression trajectory similarities of orthologous genes across different tissues and found that the expression trajectory similarity between humans and mice in the brain and cerebellum is higher than that of other tissues. This observation aligns with prior literature, which has established the brain as the most evolutionarily conserved organ during development (8). Then, we investigated a set of developmental disease genes using the DDG2P database. DDG2P database was developed to identified causative genes of developmental disorders in the DDD Study (3). The gene expression trajectory of DDG2P genes displayed more evolutionary conservation in most major organs across humans and mice than non-DDG2P genes, as evidenced by the similarity of gene expression trajectories. This finding suggested that the functional significance of DDG2P genes in development has been preserved across mammalian evolution.
The AD genes of the DDG2P database showed a significantly higher proportion of genes with similar expression trajectories in the brain and cerebellum compared to AR genes. This finding suggested that AD genes are more evolutionarily conserved than AR genes, as previously indicated (30,31). Hence, the presence of similar expression trajectories in these AD genes implies their potential role in the evolutionary conservation of brain and cerebellum development between humans and mice. Notably, this pattern consistently manifests in genes that were definitively and strongly associated with the developmental disorders. Furthermore, AD genes exert a more pivotal functional role compared to AR genes (31). Given that functional impairments of disease-causing genes contribute to developmental delay (3), the observed differential similarities in AD versus AR gene expression among genes with definitive and strong disease associations likely reflect their greater functional importance during development.
The high sequence-level similarities have made mouse models a widely used tool in scientific research (32). Additionally, as alterations in phenotypes are formed during development, the similarities of expression trajectories might appear to be important during evolution. To confirm this phenomenon, we classified the DDG2P genes based on FUSIL categorization and demonstrated that VP genes with significantly similar expression trajectories were related to more comparable phenotypes compared to genes with dissimilar trajectories. However, this conclusion cannot be generalized to other gene categories, likely due to the impact of lethal phenotypes in mice on the calculation of phenotypic similarity.
Based on the above conclusions, we focused the study on iGluRs genes family linked to neurodevelopmental disorders, in order to advance our understanding of utilizing expression trajectory similarity for predicting phenotypic similarity. Ten of the iGluRs genes had been involved in the DDG2P database and reported in the OMIM database. VP genes were the overlapped genes between genes with viable and abnormal phenotypes in knockout mouse model and human non-essential genes. At present, none of the gene families within DDG2P have been entirely categorized as VP genes. Seven of disease-causing iGluRs genes were grouped with VP bin after assessment.
The disease-causing genes of iGluRs were classified based on the similarities of expression trajectories in the brain and cerebellum, as well as phenotypic similarities. The lack of a PTV in three iGluRs genes resulted in the absence of refined Phenodigm scores, thereby leading to certain limitations. Strong agreement between high scores of phenotypic similarity and correlation coefficients of expression in the brain and cerebellum demonstrated that knockout mouse of GRIA4, GRIN1, GRIN2A and GRIN2B can accurately mimic human neurodevelopmental disorders. Differently from what was previously anticipated, genes with distinct expression profiles in the brain and cerebellum between humans and mice may result in phenotypes that are either concordant or discordant. For instance, the expression of the GRIN2D gene varies between the brain and cerebellum of humans and mice, its phenotypic score is likewise the lowest. Despite the absence of reported neurodevelopmental disorders cases associated with GRIN2D PTVs, the original Phenodigm score was employed as a reference. But GRIA1 [60], GRID2 [53] and GRIK2 [53], which displayed dissimilar expression trajectories across human and mouse, demonstrated high refined Phenodigm scores.
This observation indicated that comparable phenotypes may arise from different gene expression trajectories between humans and mice. Two potential explanations may account for this phenomenon. Firstly, genes might show similar expression trajectories in a critical period of development, rather than throughout the entire process. For instance, during early embryonic development, the expression trajectories of GRIK2 in the cerebellum of humans and mice exhibit a similar pattern (Figure S1H). However, subsequent to this period, these trajectories diverge, leading to differing patterns of gene expression between the two species. The other possible explanation is that the mechanisms underlying disease-associated genes possess the potential to align with molecular processes and pathways, revealing intricate interdependencies (33). In cases where a gene becomes dysfunctional, the remaining genes have the capacity to assume compensatory functions, thereby influencing the observed differences in phenotypes between humans and mice. Therefore, we hypothesized that analogous gene expression trajectories might be used to infer similar phenotypes, although divergent gene expression trajectories do not necessarily correspond to dissimilar phenotypes.
When genes showed different similarities of expression trajectories in the brain and cerebellum, phenotypic similarities are determined based on the similarity of expression trajectories in the organ relevant to the phenotypes. For example, GRIA2 demonstrated similar expression trajectories in the brain but not in the cerebellum. The Phenodigm score of GRIA2 obtained from matching human PTVs patients with homozygous gene deletion mouse models was the lowest. Specifically, the GRIA2 knockout mouse exhibited impaired motor coordination and anomalous behavior (26), whereas patients with PTVs had normal cerebellum imaging and motor ability (29). The long-term plasticity of synaptic transmission in the cerebellum is recognized as the key mechanism underlying motor learning. It is hypothesized that the activation of presynaptic α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor could be a molecular event involved in this fundamental process (34). Thus, the difference in motor ability between humans and mice may be attributed to the dissimilarity of gene expression trajectories in the cerebellum. The GluA2 subunits were dominant in the AMPA receptor and the expression of GRIA2 became elevated as time went on in the mouse cerebellum, while the expression gradually decreased during the mild-late embryonic development in the human cerebellum (Figure S1E). As for GRIA3, it presented similar expression trajectories in the brain of humans and mice. Although comparison of phenotypes was unable to performed due to absence of GRIA3 patients with PTVs, sleep disturbance of patients with GRIA3 missense variants was simulated in the point mutation mouse of GRIA3 (35). In rats, the transitions between wakefulness and sleep are accompanied by alterations in extracellular glutamate concentrations of cerebral cortex (36). Recent findings provide compelling evidence indicating a notable role for GluA3 channel activity in the regulation of sleep behavior in both mice and humans (35). Therefore, when the gene expression trajectories of a particular organ in the brain and cerebellum show comparable similarities between n humans and mice, it suggests that the phenotypic outcomes associated with organ-specific gene expression patterns may also exhibit similarity.
Building upon our investigation into disease-causing iGluR genes, we have demonstrated that similar expression trajectories of the brain and cerebellum between humans and mice have the potential to predict similar neurological phenotypes. Integrating the phenotypes of gene knockout mouse models might enable the aiding to predict the potential disease-causing genes.
Unknown disease-causing genes of iGluRs were subject to classification based on the similarities of their expression trajectories of the brain and cerebellum across human and mouse. The GRIK3 and GRIN2C genes displayed comparable expression trajectories in the brain and cerebellum across species. The neurological sign has not been mentioned in the GRIK3 deficient mouse models. In GRIN2C knockout mice, deficiencies in fear acquisition and working memory had been observed (37), and also provided evidence for the involvement of the GluN2C subunit in schizophrenia-like behaviors (38). Furthermore, a genomic data analysis study indicated a single nucleotide polymorphism (SNP) within GRIN2C that is associated with early-onset bipolar disorder in patients (39). An ultra-rare variant of GRIN2C may be considered to increase the risk of schizophrenia (40). These findings suggested that GRIN2C might be a potential pathogenic gene.
For genes exhibiting similarity in expression solely within the brain or cerebellum, we infer their pathogenic potential based on the associated mouse phenotypes related to organ-specific functions. GRIK4 and GRID1 genes demonstrated analogous expression trajectories in the brain between human and mouse. In mouse models, the deficiency of GRIK4 alleviated anxiety-like behavior (41), while an increase in dosage can give rise to autism or schizophrenia-related behaviors (42,43). Furthermore, a patient with autism was identified to carry a de novo duplication of the chromosome 11q23.3-q24.1 locus, which includes GRIK4 gene (44). Additionally, knockout of GRID1 gene in mice resulted in depressive-like behavior, abnormal social behavior, and impaired fear memory (45,46). Several large independent studies have also demonstrated that the GRID1 gene promoter region has common genetic variations that increase the risk of schizophrenia (47-49). Thus, duplication of GRIK4 might induce pathogenic copy number variation, and genetic variants of GRID1 might play a role in the etiology of schizophrenia.
Similar expression trajectories were detected in GRIK1 and GRIN3A in the cerebellum but not in the brain. Abnormalities of cerebellar function have not been detected in GRIK1 or GRIN3A knockout mouse, so we didn’t further discuss. In the iGluRs gene family, we have identified that similar phenotypes can be inferred based on similar expression trajectories. In combination with mouse phenotypes, this approach might be useful for predicting potential disease-causing genes. This has been validated in the instance of GRIN2C, GRIK4, and GRID1, which were identified as potential disease-causing genes.
Overall, the current work is the first to demonstrate the relationship between expression trajectory similarity and phenotypic similarity and provided a potential explanation for the distinct phenotypes resulting from null mutations in humans and mice from a novel perspective. In addition, comparing expression trajectories provides a solid basis for forthcoming investigations involving knockout mouse models. Furthermore, the use of gene expression trajectories might improve the accuracy and reliability of predicting potential pathogenic genes. This approach holds promise for advancing our understanding of gene function and disease mechanisms. Consequently, this study opens up new avenues for future research endeavors of developmental disorders.
Conclusions
In genes associated with developmental disorders, abnormal phenotypes resulting from gene mutations are more similar across species for the VP genes with similar expression in the brain of humans and mice compared to genes with dissimilar expression. By studying a gene family associated with neurodevelopmental disorders, it was found that similar neurological phenotypes could be deduced from similar gene expression trajectories in the brain and cerebellum. This approach, in conjunction with mouse phenotypes, might aid in predicting potential pathogenic genes. These findings highlight the importance of gene expression trajectories in the study of developmental disorders.
Acknowledgments
Funding: None.
Footnote
Peer Review File: Available at https://pm.amegroups.com/article/view/10.21037/pm-23-27/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://pm.amegroups.com/article/view/10.21037/pm-23-27/coif). W.Z. serves as an unpaid Executive Editor-in-Chief of Pediatric Medicine. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sheridan E, Wright J, Small N, et al. Risk factors for congenital anomaly in a multiethnic birth cohort: an analysis of the Born in Bradford study. Lancet 2013;382:1350-9. [Crossref] [PubMed]
- Ropers HH. Genetics of early onset cognitive impairment. Annu Rev Genomics Hum Genet 2010;11:161-87. [Crossref] [PubMed]
- Prevalence and architecture of de novo mutations in developmental disorders. Nature 2017;542:433-8. [Crossref] [PubMed]
- Cacheiro P, Haendel MA, Smedley D, et al. New models for human disease from the International Mouse Phenotyping Consortium. Mamm Genome 2019;30:143-50. [Crossref] [PubMed]
- Zhou Z, Tan C, Chau MHK, et al. TEDD: a database of temporal gene expression patterns during multiple developmental periods in human and model organisms. Nucleic Acids Res 2023;51:D1168-78. [Crossref] [PubMed]
- Cardoso-Moreira M, Sarropoulos I, Velten B, et al. Developmental Gene Expression Differences between Humans and Mammalian Models. Cell Rep 2020;33:108308. [Crossref] [PubMed]
- Zheng-Bradley X, Rung J, Parkinson H, et al. Large scale comparison of global gene expression patterns in human and mouse. Genome Biol 2010;11:R124. [Crossref] [PubMed]
- Cardoso-Moreira M, Halbert J, Valloton D, et al. Gene expression across mammalian organ development. Nature 2019;571:505-9. [Crossref] [PubMed]
- Köhler S, Gargano M, Matentzoglu N, et al. The Human Phenotype Ontology in 2021. Nucleic Acids Res 2021;49:D1207-17. [Crossref] [PubMed]
- Groza T, Gomez FL, Mashhadi HHThe International Mouse Phenotyping Consortium, et al. comprehensive knockout phenotyping underpinning the study of human disease. Nucleic Acids Res 2023;51:D1038-45. [Crossref] [PubMed]
- Smith CL, Goldsmith CA, Eppig JT. The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol 2005;6:R7. [Crossref] [PubMed]
- Smedley D, Oellrich A, Köhler S, et al. PhenoDigm: analyzing curated annotations to associate animal models with human diseases. Database (Oxford) 2013;2013:bat025. [Crossref] [PubMed]
- Daniel WW. Applied Nonparametric Statistics (2nd ed.). Spearman rank correlation coefficient. Boston: PWS-Kent; 1990.
- Hadley W, Mara A, Jennifer B, et al. Welcome to the Tidyverse. Journal of Open Source Software 2019;4:1686.
- Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York; 2016.
- Rivas MA, Pirinen M, Conrad DF, et al. Human genomics. Effect of predicted protein-truncating genetic variants on the human transcriptome. Science 2015;348:666-9. [Crossref] [PubMed]
- Köhler S, Carmody L, Vasilevsky N, et al. Expansion of the Human Phenotype Ontology (HPO) knowledge base and resources. Nucleic Acids Res 2019;47:D1018-27. [Crossref] [PubMed]
- Smith CL, Blake JA, Kadin JA, et al. Mouse Genome Database (MGD)-2018: knowledgebase for the laboratory mouse. Nucleic Acids Res 2018;46:D836-42. [Crossref] [PubMed]
- Smith CL, Eppig JT. The mammalian phenotype ontology: enabling robust annotation and comparative analysis. Wiley Interdiscip Rev Syst Biol Med 2009;1:390-9. [Crossref] [PubMed]
- Thormann A, Halachev M, McLaren W, et al. Flexible and scalable diagnostic filtering of genomic variants using G2P with Ensembl VEP. Nat Commun 2019;10:2373. [Crossref] [PubMed]
- Cacheiro P, Muñoz-Fuentes V, Murray SA, et al. Human and mouse essentiality screens as a resource for disease gene discovery. Nat Commun 2020;11:655. [Crossref] [PubMed]
- Kassambara A. rstatix: Pipe-Friendly Framework for Basic Statistical Tests. 2021. Available online: https://CRAN.R-project.org/package=rstatix. Accessed December 11 2022.
- Team RC. R: A Language Environment for Statistical Computing. 2020. Available online: https://www.R-project.org/. Accessed December 11 2022.
- Stroebel D, Paoletti P. Architecture and function of NMDA receptors: an evolutionary perspective. J Physiol 2021;599:2615-38. [Crossref] [PubMed]
- Parenti I, Rabaneda LG, Schoen H, et al. Neurodevelopmental Disorders: From Genetics to Functional Pathways. Trends Neurosci 2020;43:608-21. [Crossref] [PubMed]
- Jia Z, Agopyan N, Miu P, et al. Enhanced LTP in mice deficient in the AMPA receptor GluR2. Neuron 1996;17:945-56. [Crossref] [PubMed]
- Li Y, Erzurumlu RS, Chen C, et al. Whisker-related neuronal patterns fail to develop in the trigeminal brainstem nuclei of NMDAR1 knockout mice. Cell 1994;76:427-37. [Crossref] [PubMed]
- Sprengel R, Suchanek B, Amico C, et al. Importance of the intracellular domain of NR2 subunits for NMDA receptor function in vivo. Cell 1998;92:279-89. [Crossref] [PubMed]
- Salpietro V, Dixon CL, Guo H, et al. AMPA receptor GluA2 subunit defects are a cause of neurodevelopmental disorders. Nat Commun 2019;10:3094. [Crossref] [PubMed]
- Furney SJ, Albà MM, López-Bigas N. Differences in the evolutionary history of disease genes affected by dominant or recessive mutations. BMC Genomics 2006;7:165. [Crossref] [PubMed]
- Hao D, Wang G, Yin Z, et al. Systematic large-scale study of the inheritance mode of Mendelian disorders provides new insight into human diseasome. Eur J Hum Genet 2014;22:1260-7. [Crossref] [PubMed]
- Waterston RH, Lindblad-Toh K, et al. Initial sequencing and comparative analysis of the mouse genome. Nature 2002;420:520-62. [Crossref] [PubMed]
- Tan AP, Chong WK, Mankad K. Comprehensive genotype-phenotype correlation in lissencephaly. Quant Imaging Med Surg 2018;8:673-93. [Crossref] [PubMed]
- Zanetti L, Regoni M, Ratti E, et al. Presynaptic AMPA Receptors in Health and Disease. Cells 2021;10:2260. [Crossref] [PubMed]
- Davies B, Brown LA, Cais O, et al. A point mutation in the ion conduction pore of AMPA receptor GRIA3 causes dramatically perturbed sleep patterns as well as intellectual disability. Hum Mol Genet 2017;26:3869-82. [Crossref] [PubMed]
- Dash MB, Douglas CL, Vyazovskiy VV, et al. Long-term homeostasis of extracellular glutamate in the rat cerebral cortex across sleep and waking states. J Neurosci 2009;29:620-9. [Crossref] [PubMed]
- Hillman BG, Gupta SC, Stairs DJ, et al. Behavioral analysis of NR2C knockout mouse reveals deficit in acquisition of conditioned fear and working memory. Neurobiol Learn Mem 2011;95:404-14. [Crossref] [PubMed]
- Shelkar GP, Pavuluri R, Gandhi PJ, et al. Differential effect of NMDA receptor GluN2C and GluN2D subunit ablation on behavior and channel blocker-induced schizophrenia phenotypes. Sci Rep 2019;9:7572. [Crossref] [PubMed]
- Forero DA, Herteleer L, De Zutter S, et al. A network of synaptic genes associated with schizophrenia and bipolar disorder. Schizophr Res 2016;172:68-74. [Crossref] [PubMed]
- Yu Y, Lin Y, Takasaki Y, et al. Rare loss of function mutations in N-methyl-D-aspartate glutamate receptors and their contributions to schizophrenia susceptibility. Transl Psychiatry 2018;8:12. [Crossref] [PubMed]
- Mennesson M, Rydgren E, Lipina T, et al. Kainate receptor auxiliary subunit NETO2 is required for normal fear expression and extinction. Neuropsychopharmacology 2019;44:1855-66. [Crossref] [PubMed]
- Arora V, Pecoraro V, Aller MI, et al. Increased Grik4 Gene Dosage Causes Imbalanced Circuit Output and Human Disease-Related Behaviors. Cell Rep 2018;23:3827-38. [Crossref] [PubMed]
- Aller MI, Pecoraro V, Paternain AV, et al. Increased Dosage of High-Affinity Kainate Receptor Gene grik4 Alters Synaptic Transmission and Reproduces Autism Spectrum Disorders Features. J Neurosci 2015;35:13619-28.
- Griswold AJ, Ma D, Cukier HN, et al. Evaluation of copy number variations reveals novel candidate genes in autism spectrum disorder-associated pathways. Hum Mol Genet 2012;21:3513-23. [Crossref] [PubMed]
- Yadav R, Gupta SC, Hillman BG, et al. Deletion of glutamate delta-1 receptor in mouse leads to aberrant emotional and social behaviors. PLoS One 2012;7:e32969. [Crossref] [PubMed]
- Nakamoto C, Kawamura M, Nakatsukasa E, et al. GluD1 knockout mice with a pure C57BL/6N background show impaired fear memory, social interaction, and enhanced depressive-like behavior. PLoS One 2020;15:e0229288. [Crossref] [PubMed]
- Treutlein J, Mühleisen TW, Frank J, et al. Dissection of phenotype reveals possible association between schizophrenia and Glutamate Receptor Delta 1 (GRID1) gene promoter. Schizophr Res 2009;111:123-30. [Crossref] [PubMed]
- Fallin MD, Lasseter VK, Avramopoulos D, et al. Bipolar I disorder and schizophrenia: a 440-single-nucleotide polymorphism screen of 64 candidate genes among Ashkenazi Jewish case-parent trios. Am J Hum Genet 2005;77:918-36. [Crossref] [PubMed]
- Chen X, Lee G, Maher BS, et al. GWA study data mining and independent replication identify cardiomyopathy-associated 5 (CMYA5) as a risk gene for schizophrenia. Mol Psychiatry 2011;16:1117-29. [Crossref] [PubMed]
Cite this article as: Jiang X, Tao J, Zhang Y, Lin Z, Wang Y, Zhou W. Gene expression trajectories aiding the unveiling of phenotypic similarities in developmental disorders: insights from human-mouse comparative analysis. Pediatr Med 2023;6:32.

