
Review of the contribution of genetic factors to hyperbilirubinemia and kernicterus risk in neonates: a targeted update
Introduction
The genesis of significant neonatal hyperbilirubinemia [total serum bilirubin (TSB) ≥17 mg/dL (291 µmol/L)] (1) is characterized by etiologic heterogeneity, environmental modulation, and the interaction of multiple gene loci (2-6). Comprehensive reviews of specific genetic contributors to neonatal jaundice have been published and suggest that in addition to inherited hemolytic conditions such as hereditary spherocytosis, common icterogenic gene variants with individually small effects may act as modifiers of hyperbilirubinemia and kernicterus risk (2-6). Damaging mutations (e.g., those of Crigler-Najjar type I) also contribute to the overall genetic architecture of neonatal hyperbilirubinemia but, fortunately, are rare. The current review targets the effect biologic sex, uridine diphosphate glucuronosyltransferase isoenzyme UGT1A1 (OMIM *191740) gene variants of Gilbert syndrome (OMIM #143500), and co-expression of icterogenic alleles have on potentiating hyperbilirubinemia risk in neonates.
Sexual dimorphism in neonatal hyperbilirubinemia and kernicterus risk
One of the most frequently reported, yet often overlooked, genetic based contributors to hyperbilirubinemia and kernicterus risk is the biologic sex of the neonate. Male neonates have higher TSB concentrations (7-9), higher rates of non-physiologic hyperbilirubinemia [TSB >12 mg/dL; >205 µmol/L (10)] and are at greater risk for hospital readmission for neonatal jaundice (9,11,12) than female neonates. Similarly, a male sex bias typifies extreme (TSB ≥25 mg/dL) and hazardous (TSB ≥30 mg/dL) hyperbilirubinemia cohorts (13) as well as kernicterus registries (1,14-20). The greater male susceptibility to bilirubin-induced brain damage, on an order of more than 2:1 in some reports (1,14,20), is a consistent finding across many countries, including the United States (1,14), the United Kingdom and Ireland (15), Canada (16), Egypt (17), Denmark (19), Sweden (18), and China (20). Earlier studies reported a male preponderance in preterm kernicterus (21), in neonatal mortality attributed to kernicterus in erythroblastosis fetalis (22), and correspondingly in autopsy case series of kernicterus (23).
Glucose-6-phosphate dehydrogenase (G6PD) deficiency, an X-linked condition consistently overrepresented as a cause of kernicterus across the globe (24), in all probability contributes to the male preponderance in bilirubin-induced brain damage. Similarly, Gilbert syndrome, a hyperbilirubinemia potentiating genetic condition, is more prevalent in males (25-28). It is doubtful, however, that these clinical entities alone account for the greater numbers of affected males among kernicterus cases.
Although recognition of this male sex bias has no relevance to the care of neonates (female neonates are at risk for kernicterus and are evaluated and managed the same as their male counterparts), it is surprising the male sex kernicterus bias has received limited examination and not been exploited to enhance our understanding of bilirubin-induced brain injury.
The Gunn rat model of neonatal hyperbilirubinemia mirrors humans in sharing a male sex bias for kernicterus and thereby lends itself as a tool for investigating the nature of sexual dimorphism in kernicterus (29,30). Existing data on male-female differences in jaundiced (j/j) hyperbilirubinemic Gunn rat pups, including those collected during sulfadimethoxine induced acute bilirubin encephalopathy, are limited and shown in Table 1. They demonstrate that advanced stages of neuromotor dysfunction and kernicterus in hyperbilirubinemic male j/j Gunn rat pups are associated with a two-fold greater cerebellar and brainstem bilirubin content than in their jaundiced female j/j littermate pairs (30). Given similar baseline TSB, serum albumin, and calculated free bilirubin levels (30), one would presume the sulfadimethoxine induced CNS bilirubin exposure itself should be similar between male and female j/j Gunn rat pups. The notable greater cerebellar bilirubin content in j/j male pups may therefore reflect sex specific differences in CNS bilirubin uptake and clearance (30). CNS bilirubin content is modulated in part by the efflux transporter P-glycoprotein (33-35) an ATP binding cassette (ABC) transmembrane protein encoded by the ABCB1 gene and expressed in microvessel endothelial cells of the blood-brain barrier in human neonates (36). There are, however, no data on sex specific CNS P-glycoprotein expression patterns or the impact of non-synonymous ABCB1 gene variants (37,38) on P-glycoprotein function in the neonatal period.
Table 1
| Variables | Male | Female | Reference |
|---|---|---|---|
| % BIND score* ≥2 | ~80% | ~50% | Unpublished observations |
| Total serum bilirubin (mg/dL)** | 7.1±1.2 | 7.5±1.1 | (30) |
| Serum albumin (g/dL)** | 3.2±0.6 | 3.1±0.5 | (30) |
| Calculated unbound bilirubin (µmol/L)** | 0.149±0.028 | 0.153±0.021 | (30) |
| Cerebellar bilirubin content (ug/g tissue) | 17.9±8.8# | 9.2±6.8 | (30) |
| Brainstem bilirubin content (ug/g tissue) | 10.8±8.1 | 6.8±2.9 | (30) |
| Microglia (per hpf)+ | 39.8±27.8 | 18.9±7.4 | Unpublished observations |
| Activated microglia (per hpf)+ | 17.0±11.2 | 10.5±8.2 | Unpublished observations |
| Activated microglia in granular layer (per hpf)+ | 14.0±5.8 | 6.7±6.1 | Unpublished observations |
| Kernicterus† | 57.6% | 40.5% | (29) |
Except where otherwise indicated, data were measured 24 hours following sulfadimethoxine triggered acute bilirubin encephalopathy. *, bilirubin-induced neurologic dysfunction (BIND) score quantifies gait abnormalities and dystonia in hyperbilirubinemic j/j Gunn rats. Score ranges from 0 to 5 based on the following signs: 0= normal; 1= mildly abnormal with slight hindlimb ataxia; 2= mild hindlimb ataxia, dystonia and gait abnormality with impaired righting reflex; 3= abnormal as in 2, but with more severe movement disorder and prolonged righting reflex; 4= severe failure of locomotion, general lack of spontaneous movement with occasional bursts of hyperactivity and no righting reflex; 5= moribund including seizures and/or agonal respirations (31,32). **, prior to sulfadimethoxine dosing. #, P<0.02 compared with female cerebellum (Cannon 2006). +, cerebellar microglia CD11b/c immunofluorescence (OX-42, Serotec, Raleigh, NC) counts were characterized as activated microglia if they displayed a larger, more rounded amoeboid soma and thicker less ramified processes. Counts were expressed as number of microglia per high powered field (400×) based on a minimum of 3 non-overlapping fields (mean ± SD). †, in absence of sulfadimethoxine induced bilirubin encephalopathy.
Table 1 also highlights a more robust microglial response in j/j male pups during sulfadimethoxine induced acute bilirubin encephalopathy. Although neuroinflammation routinely accompanies bilirubin induced brain damage (39), it is unclear if the greater number of microglia, including amoeboid activated microglia, in encephalopathic male j/j Gunn rat pup cerebellum simply mirrors or contributes to the bilirubin-induced injury.
One potential experimental approach to explore male-female j/j Gunn rat pup differences in susceptibility to bilirubin-induced brain injury is hormonal manipulation. Plasma estradiol in the female Gunn rat pups is protein bound and does not exert an effect in the CNS. By contrast, males produce testosterone which is able to cross the BBB and enter the CNS where it either exerts a direct androgenic effect or is converted to estradiol by tissue aromatase in a region-specific fashion (40). Notably, aromatase levels are undetectable in both male and female rat pup cerebellum (41,42) including the Gunn rat strain (unpublished observations). Other possible experimental manipulations could include castrating Gunn rat male pups or treating females with testosterone. Regardless, the male sex bias in bilirubin-induced brain injury merits further study in the continued effort to more fully understand the cascade of events leading to kernicterus.
Gilbert syndrome and the neonate
Gilbert syndrome is a common congenital inborn error of hepatic bilirubin conjugation wherein UGT1A1 isoenzyme activity is reduced by ~70% or more (43,44). Many pediatricians have long speculated a role for Gilbert syndrome in potentiating neonatal hyperbilirubinemia (45-48). Following the seminal genetic characterizations of Gilbert syndrome (43,49-51) more than twenty-five years ago, support for this conjecture has grown as has our understanding of the roles genetic heterogeneity and UGT1A1 variant allele co-expression play in this condition.
Molecular genetics of Gilbert syndrome
A schematic diagram of the UGT1A1 gene is shown in Figure 1. Originally defined by an extra thymine-adenine (TA) dinucleotide repeat within the A(TA)nTAA element of the UGT1A1 TATAA box promoter (UGT1A1*28) in European populations (43,49) and missense mutations in UGT1A1 exons (UGT1A1*6, UGT1A1*7, UGT1A1*27, UGT1A1*29) in Japan (50,51), at least 14 additional UGT1A1 variant alleles have been identified in association with a Gilbert syndrome phenotype (Table 2). Generally held to be an autosomal recessive condition (63), Gilbert syndrome may be inherited in an autosomal dominant manner when UGT1A1 exon variants are operative; UGT1A1*6 is a prime example (50,51,64-66). Inheritance is subject to variable penetrance and expressivity (43) depending on the nature of the UGT1A1 variant, the co-expression of modifying alleles, and the presence of environmental factors.
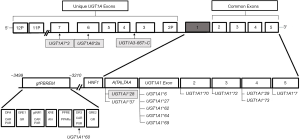
Table 2
| Allele | Nucleotide change | Amino acid change | Variant location | Reference |
|---|---|---|---|---|
| UGT1A1*1 | A(TA)6TAA | Wild type | Promoter | – |
| UGT1A1*6 | 211(G>A) | G71R | Exon 1 | (54) |
| UGT1A1*28 | A(TA)6TAA to A(TA)7TAA | n/a | Promoter | (43) |
| UGT1A1*60 | −3279(T>G) | n/a | Promoter | (55,56) |
| UGT1A1*7 | 1456(T>G) | Y486D | Exon 5 | (54) |
| UGT1A1*27 | 686(C>A) | P229Q | Exon 1 | (51) |
| UGT1A1*37 | A(TA)6TAA to A(TA)8TAA | n/a | Promoter | (57) |
| UGT1A1*62 | 247(T>C) | F83L | Exon 1 | (58) |
| UGT1A1*64 | 488-491 dupACCT | Frameshift | Exon 1 | (59) |
| UGT1A1*65 | −1126(C>T) | n/a | Promoter | (59) |
| UGT1A1*66 | 997-82(T>C) | n/a | Intron 2 | (59) |
| UGT1A1*67 | −85 to −83 ins CAT | n/a | Promoter | (60) |
| UGT1A1*68 | −63(G>C) | n/a | Promoter | (60) |
| UGT1A1*69 | 476(T>C) | I159T | Exon 1 | (60) |
| UGT1A1*70 | 962(C>G) | A321G | Exon 2 | (60) |
| UGT1A1*72 | 1075(G>A) | D359N | Exon 3 | (60) |
| UGT1A1*73 | 1091(C>T) | P364L | Exon 4 | (60) |
| UGT1A1*81 | −64(G>C) | n/a | Promoter | (61) |
Alleles shaded in gray are polymorphic. Adapted updated and modified from reference (62). Reproduced with permission of Taylor & Francis Inc.
Adding to this complexity, Ehmer et al. report that Gilbert syndrome often represents an expanded genetic haplotype encompassing co-expression of UGT1A3, UGT1A6, and UGT1A7 variants in addition to those of UGT1A1 (Figure 1) (53). Greater than three quarters of individuals homozygous for UGT1A1*28 from the Ehmer et al. white northern European cohort were concurrently homozygous for UGT1A3-66 T>C, UGT1A6*2a and UGT1A7*3 (53). Moreover, higher TSB levels were observed in those carrying the expanded four gene UGT1A haplotype than in those homozygous for UGT1A1*28 alone (53). It is unclear, however, whether this or an analogous expanded haplotype is expressed in other populations. Similarly, it is uncertain how the expanded four gene UGT1A haplotype enhances hyperbilirubinemia given that only UGT1A1 effectively conjugates bilirubin (67). Nothing is known about the perinatal, neonatal or postnatal expression of this expanded UGT1A haplotype or how it might impact neonatal hyperbilirubinemia risk. Regardless, this review will only examine UGT1A1 variant polymorphisms of Gilbert syndrome.
Gilbert syndrome and neonatal hyperbilirubinemia risk
Bancroft et al. in 1998 were the first to explore the relationship between a Gilbert syndrome genotype, specifically UGT1A1*28, and neonatal hyperbilirubinemia (68). Their findings, from a largely white-non-Hispanic cohort, demonstrated an increased rate of rise in transcutaneous bilirubin levels during the first two days of life in UGT1A1*28 homozygous neonates (68). Despite the enhanced rate of rise, peak transcutaneous bilirubin levels in neonates with Gilbert syndrome did not differ from wild type controls (68). Bancroft et al. concluded that the “determination of the relative role of this genetic variable in the assessment of overall neonatal jaundice risk will require completion of a prospective study with multivariate analysis to examine various combinations of jaundice risk factors” (68).
Numerous such studies published over the ensuing decades support a potentiating role for Gilbert syndrome in neonatal hyperbilirubinemia risk, depending on the specific genotype, study population, the presence of breastmilk feeding, and/or hemolytic disease. The most prevalent polymorphic gene variants involved are UGT1A1*28, UGT1A1*6, and UGT1A1*60.
UGT1A1*28
The Gilbert syndrome promoter sequence variant UGT1A1*28 is common to individuals of European and African ancestry (57,69,70). Several studies (2,3,64,71-76), but not all (10,65,77,78), suggest that UGT1A1*28 alone poses limited to no enhanced neonatal hyperbilirubinemia risk. Published meta-analyses confirm the same (64) or at least less icterogenic potential than UGT1A1*6 (64). The UGT1A1*28 variant, however, when co-expressed with UGT1A1 coding sequence variants or icterogenic conditions such as breastfeeding and hemolytic disease, appears to augment hyperbilirubinemia risk.
UGT1A1*6
In marked contrast to UGT1A1*28, it is increasingly apparent that the UGT1A1 exon 1 coding sequence variant UGT1A1*6, common to Asian populations, in itself exerts an icterogenic effect. UGT1A1*6 acts as an independent risk factor for neonatal hyperbilirubinemia (64-66,79-82) and contributes to, and in fact may primarily underlie, the widely recognized increased neonatal hyperbilirubinemia risk among Asian populations (83-85). This association is robust as confirmed by at least three large meta-analyses, irrespective of genetic modeling approach (homozygous, heterozygous, dominant or recessive) (64-66), a fundamental effect that may relate to the greater reduction in UGT1A1 enzymatic activity (~14% of wild type levels) associated with this variant (50). Not surprisingly, co-expression of UGT1A1*6 with other coding sequence variants, promoter variants, hemolytic conditions, and breastfeeding further increases hyperbilirubinemia risk.
UGT1A1*60
The UGT1A1*60 variant has been of great interest in efforts to understand the genetic nature of Gilbert syndrome. This promoter variant is located in the glucuronosyltransferase phenobarbital responsive enhancer module (gtPBREM) cluster (Figure 1), a regulatory element containing multiple binding sites for nuclear receptor motifs including the constitutive androstane receptor (CAR), the pregnane X receptor (PXR), the xenobiotic responsive element (XRE), peroxisome proliferator-activated receptor alpha (PPARα), and glucocorticoid responsive elements (GRE) (52). UGT1A1*60, in contrast to most other variants, is common across studied populations regardless of biogeographic heritage with allele frequencies generally between 0.25 and 0.50 (3). Studies report a high degree of linkage disequilibrium between UGT1A1*60 and UGT1A1*28, an association asserted by some (69) [but not all (86,87)] as essential to the genesis of Gilbert syndrome. Linkage of UGT1A1*60 with UGT1A1*6 has also been reported (28). However, investigations suggest that homozygous expression of UGT1A1*60 alone can be associated with reduced UGT1A1 transcriptional activity (55,56) and itself account for Gilbert syndrome in some populations (56). A recent meta-analysis shows that UGT1A1*60 is associated with a significant increased risk for neonatal hyperbilirubinemia, albeit the study did not rule out linkage with other variants as the mechanism (88). Given the cluster of regulatory elements contained in gtPBREM and their potential roles in regulating the developmental expression of UGT1A1, it is important to further clarify the nature of variants localized to that region of the UGT1A1 promoter (52,89,90).
Spectrum of neonatal hyperbilirubinemia risk in Gilbert syndrome
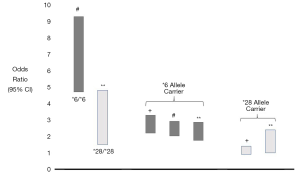
Taken together the UGT1A1*28 and UGT1A1*6 studies detailed above suggest there is a spectrum of neonatal hyperbilirubinemia risk across Gilbert syndrome genotypes (64-66), depending on the variant(s) involved and expression mode. Neonates homozygous for UGT1A1*6 demonstrate the highest reported odds ratios for neonatal hyperbilirubinemia risk among Gilbert syndrome variants (65,66), one that is notably higher than those homozygous for UGT1A1*28 (Figure 2). UGT1A1*6 allele carriers also evidence a significantly increased neonatal hyperbilirubinemia risk compared to wild type (64-66) whereas allele carriers for UGT1A1*28 show borderline to non-significant associations with neonatal hyperbilirubinemia risk (64,65) (Figure 2). Monaghan et al. suggested a similar divergence in hyperbilirubinemia risk in their 1996 study of adults when they asserted there were both “mild and more severe forms of Gilbert’s syndrome” (49). The ‘mild’ form was related to expression of UGT1A1 promoter variants whereas ‘more severe forms’ were due to UGT1A1 coding sequence variants (49). In fact, promoter variants alone may not always be sufficient to develop a Gilbert syndrome phenotype (43), whereas biallelic expression of coding sequence variants associated with Gilbert syndrome may lead to TSB levels intermediate between Gilbert syndrome and Crigler-Najjar syndrome type II (91). Indeed, the genetic characterization of the non-hemolytic hyperbilirubinemias Gilbert syndrome, Crigler-Najjar syndrome type II and Crigler-Najjar syndrome type I are not as invariant and sharply demarcated as once thought, but similarly reflect a spectrum of hyperbilirubinemia severity and risk.
UGT1A1 gene variants and hemolysis
Hemolysis is the dominant cause of extreme hyperbilirubinemia, acute bilirubin encephalopathy, and kernicterus (92,93). Expression of UGT1A1 gene variants of Gilbert syndrome with hemolytic conditions may augment the risk of significant hyperbilirubinemia (2-5,24,94,95). Kaplan et al. was the first to highlight the importance of this phenomenon in G6PD deficiency, itself a leading cause of kernicterus worldwide (94). In their seminal study, there was a dose dependent genetic interaction of UGT1A1*28 alleles in hemizygous G6PD Mediterranean deficient males that enhanced neonatal hyperbilirubinemia (TSB >15 mg/dL) risk (94). A similar association has been reported between UGT1A1*6 and G6PD deficiency in China (95). Others have documented an association between UGT1A1*28 and hyperbilirubinemia risk in symptomatic ABO hemolytic disease of the newborn, hereditary spherocytosis, G6PD deficiency, and beta-thalassemia (96-100). Correspondingly, a recent report from China demonstrates co-expression of UGT1A1*6 and ANK1 mutations of hereditary spherocytosis (101). The Gilbert syndrome variants icterogenic augmenting effect in hemolytic conditions does not appear to be related to any change in heme catabolism (96). Collectively, these studies illustrate the importance of coupling genetically determined hemolytic conditions with gene polymorphisms that reduce hepatic bilirubin clearance in increasing the risk of developing severe neonatal hyperbilirubinemia.
Breastmilk jaundice: a prevalent Gilbert syndrome phenotype
Neonatal hyperbilirubinemia is more common and TSB levels are significantly higher in breastfed than in formula-fed neonates (102,103). Hyperbilirubinemia in association with suboptimal breastmilk intake during the first week of life is termed “breast-feeding jaundice”; whereas prolonged jaundice in thriving breastfed neonates extending into the second to third week of life, occasionally longer, is termed “breastmilk jaundice” or “breastmilk jaundice syndrome” (102,103). Despite decades of investigation, the operative mechanism(s) underlying breastmilk jaundice syndrome remain a source of debate; the condition is likely multifactorial in nature.
Clinical evidence accrued during the past two decades, from around the globe, confirm that breastmilk jaundice is a prevalent Gilbert syndrome phenotype in neonates (10,104-106). Monaghan et al. were the first to highlight the association between UGT1A1 gene variants and prolonged unconjugated hyperbilirubinemia in breastfed term neonates (104). Their study of breastfed Scottish neonates showed that those homozygous for the Gilbert syndrome UGT1A1*28 promoter variant had a more than four-fold increased rate (27%) of prolonged jaundice (TSB >150 µmol/L at 14 day) than breast fed infants who were homozygous for the wild type UGT1A1*1 allele (6%) (104). Zaja et al. demonstrated a similar four-fold impact of homozygous UGT1A1*28 expression on the risk of breastmilk jaundice of greater than 21 days duration in their large Croatian cohort (10). Forty percent were homozygous for UGT1A1*28 Gilbert genotype (10).
Maruo et al. demonstrated an analogous relationship between breastmilk jaundice and the UGT1A1*6 Gilbert variant in Japan (105). They reported that 16 of 17 breastfed Japanese infants with prolonged unconjugated hyperbilirubinemia [TSB >10 mg/dL (171 µmol/L) at 3–4 weeks of age] carried at least one UGT1A1*6 variant allele. Nine of the 16 were either homozygous for UGT1A1*6 (n=8) or compound heterozygous for UGT1A1*6 and UGT1A1*28 (n=1), both classic Gilbert syndrome genotypes. The homozygous subset evidenced a median TSB of 18.8 mg/dL (range, 10.3–31.8 mg/dL) at 3–4 weeks of age. In a subsequent expanded set of 170 infants with breastmilk jaundice syndrome, Maruo et al. observed that 88 (51.8%) were homozygous for UGT1A1*6, as opposed to none in breastfed controls without breast milk jaundice (106). They also reported 23 neonates who were compound heterozygous for Gilbert variant alleles, bringing to a total of 122 (72%) the number of breastmilk jaundice infants who carried a Gilbert genotype. If one further considers heterozygosity for UGT1A1*6 a Gilbert genotype, a widely held premise, then another 26 breastmilk jaundice neonates would be added to the total, resulting in 148 of the 170-breastmilk jaundice cohort (87%) carrying a Gilbert syndrome genotype (106).
Several other studies have demonstrated a strong association between the risk (80,81), degree (79,80,107), and duration (107) of hyperbilirubinemia and a Gilbert syndrome genotype in breastfed neonates from China, Taiwan, and Greece. Other findings from these studies include a synergistic effect of UGT1A1*6 and breastfeeding on the risk of meriting phototherapy (81) and developing a TSB of ≥20 mg/dL (342 µmol/L) (79).
How does breastmilk combine with Gilbert syndrome to produce prolonged indirect hyperbilirubinemia? Ramos et al. hypothesized that both a breastmilk inhibitor of bilirubin conjugation and an impaired hepatic bilirubin conjugating system are required for the clinical expression of breastmilk jaundice syndrome (108). This conjecture is consistent with the fact that not all breastfed neonates develop breast milk jaundice syndrome and that formula-fed infants with Gilbert syndrome do not evidence prolonged indirect hyperbilirubinemia. Several different substances in breast milk have been suggested to inhibit hepatic bilirubin conjugation including pregnane-3α,20β-diol, nonesterified fatty acids, and oligosaccharides. Studies by Tukey and colleagues using a translational hyperbilirubinemic humanized UGT1 mouse model suggest an important repressive effect of breastmilk oligosaccharides on intestinal (as opposed to hepatic) UGT1A1 expression in driving breast milk jaundice risk (90,109,110). More specifically, human milk oligosaccharides block intestinal Toll-like receptor activation and downstream IĸB kinase phosphorylation (90,109,110). This in turn represses newborn intestinal UGT1A1 activity (90). Formula feeding, by contrast, activates IĸB and induces intestinal (but not hepatic) UGT1A1 activity thereby lowering the TSB (90). Whether this phenomenon is operative in human neonates is unclear. UGT1A1 is expressed in adult small intestine (duodenum, jejunum and ileum) (111,112), stomach, and colon (112) where it is localized to the epithelial cell layer of the mucosa, most prominently at the apical portion of the crypt enterocytes (111). There are, however, no comparable developmental data on intestinal UGT1A1 expression in the human fetus or neonate, a knowledge gap that is ripe for clinical investigation.
It is also possible that breastmilk and Gilbert syndrome exert their effects via the enterohepatic circulation of bilirubin to produce breastmilk jaundice. Breastmilk increases intestinal bilirubin absorption (113,114) independent of any augmentation of intestinal beta-glucuronidase activity. An important feature of Gilbert syndrome is a predominance of bilirubin monoglucuronides over bilirubin diglucuronides (115,116). This condition should increase enterohepatic bilirubin circulation as hydrolysis of monoglucuronides back to unconjugated bilirubin occurs at rates 4–6 time that of the diglucuronide (117). Combined, these breastmilk and Gilbert syndrome enterohepatic circulation enhancing effects would increase the hepatic bilirubin load while at the same time limit the liver’s capacity to conjugate that load, producing an increased prolonged unconjugated hyperbilirubinemia risk.
Biogeographic distribution of UGT1A1 Gilbert syndrome variants
As detailed above, the most prevalent genotype underlying Gilbert syndrome in European populations is the TATA box promoter variant UGT1A1*28 (3,43,69,118). Similarly, UGT1A1*28 underlies Gilbert syndrome in Sub-Saharan Africa and individuals of African biogeographic heritage where the less frequent UGT1A1*37 promoter variant allele is also observed (57,69,119). Coding sequence missense variants associated with Gilbert syndrome (e.g., UGT1A1*6) are distinctly uncommon in Northern European or African populations (3,69).
In marked contrast, UGT1A1*6 is the most common variant underlying Gilbert syndrome across many Asian populations, whereas UGT1A1*28 is less common with the exception being populations of the Indian subcontinent (India, Bangladesh, Sri Lanka) (77,119). Not surprisingly, subgroup analyses by East Asian ethnicity document that UGT1A1*6 allele carriers have a significantly increased risk of neonatal hyperbilirubinemia in Northeast Asia, Southeast Asia, China, Japan, and Malaysia (3,64,82).
The nature of these biogeographic differences in Gilbert syndrome genotypes remains a focus of investigation; but distinct genotypes leading to a Gilbert syndrome phenotype is consistent with convergent evolution (120). Correspondingly, investigators postulate that UGT1A1 variant alleles represent balanced polymorphisms in human evolution maintained by natural selection (57,69). Whether this is the case is uncertain (119), as is whether bilirubin is the source of selective pressure (69,120,121). There is growing evidence, however, that a mildly elevated TSB, an endogenous antioxidant, is associated with relative protection against an array of diseases (120,122) and may thereby provide an evolutionary advantage (3,57,69,120,121).
Co-expression of icterogenic gene polymorphisms
Co-expression of gene polymorphisms that potentiate bilirubin production, limit hepatic bilirubin uptake, reduce hepatic bilirubin conjugation and clearance is common (2,3,123) and contributes to neonatal hyperbilirubinemia risk (2,3,79,124). Such co-expression includes two interesting and clinically relevant phenomena: compound and synergistic heterozygosity. Compound heterozygosity refers to the inheritance of alternate alleles from each parent located at different loci within the same gene. A prime example is compound heterozygosity for UGT1A1 variants of Gilbert syndrome, a phenomenon more frequent than previously thought (Table 3) (27). Sun et al. recently reported as many as three and four UGT1A1 mutation sites in individuals with Gilbert syndrome (Table 3) (27).
Table 3
| Co-expressed UGT1A1 variant alleles | Allele location | Ref |
|---|---|---|
| UGT1A1*6/UGT1A1*7 | Exon 1/exon 5 | (27) |
| UGT1A1*6/UGT1A1*27 | Exon 1/exon1 | (91) |
| UGT1A1*6/UGT1A1*28 | Exon 1/promoter | (27) |
| UGT1A1*6/UGT1A1*60 | Exon 1/enhancer | (27) |
| UGT1A1*6/UGT1A1*73 | Exon 1/exon 4 | (125) |
| UGT1A1*7/UGT1A1*73 | Exon 1/exon 4 | (125) |
| UGT1A1*27/UGT1A1*60 | Exon 1/enhancer | (91) |
| UGT1A1*28/UGT1A1*7 | Promoter/exon 5 | (27) |
| UGT1A1*28/UGT1A1*27 | Promoter/exon 1 | (125) |
| UGT1A1*28/UGT1A1*29 | Promoter/exon 4 | (91) |
| UGT1A1*28/UGT1A1*60 | Promoter/enhancer | (27) |
| UGT1A1*28/UGT1A1*73 | Promoter/exon 4 | (125) |
| UGT1A1*60/UGT1A1*81 | Enhancer/promoter | (28) |
| UGT1A1*6/UGT1A1*27/UGT1A1*28 | Exon 1/exon 1/promoter | (125) |
| UGT1A1*6/UGT1A1*28/UGT1A1*60 | Exon 1/promoter/enhancer | (27) |
| UGT1A1*6/UGT1A1*28/UGT1A1*73 | Exon 1/promoter/exon 4 | (125) |
| UGT1A1*6/UGT1A1*60/UGT1A1*73 | Exon 1/enhancer/exon 4 | (27) |
| UGT1A1*28/UGT1A1*27/UGT1A1*60 | Promoter/exon1/enhancer | (27) |
| UGT1A1*28/UGT1A1*60/UGT1A1*73 | Promoter/enhancer/exon 4 | (27) |
| UGT1A1*6/UGT1A1*27/UGT1A1*28/UGT1A1*60 | Exon 1/exon 1/promoter/enhancer | (27) |
| UGT1A1*28/UGT1A1*27/UGT1A1*60/UGT1A1*73 | Promoter/exon1/enhancer/exon 4 | (27) |
In contrast, synergistic heterozygosity refers to heterozygosities across different genes that combine to produce a range of subtle to more severe phenotypes (126). Although initially applied to inborn errors of energy metabolism, synergistic heterozygosity is a concept with broad clinical applicability including the numerous genes involved in bilirubin production, metabolism, and clearance (3). Partial defects in one or more of these pathways may contribute to hyperbilirubinemia risk (3). Zangen and co-workers described the occurrence of fatal kernicterus in a female neonate heterozygous for the G6PD Mediterranean mutation and the UGT1A1*28 Gilbert syndrome variant as a paradigm for this phenomenon in neonatal hyperbilirubinemia (127).
Next generation sequencing in the diagnostic evaluation of extreme or hazardous neonatal hyperbilirubinemia and kernicterus
In almost all reported case series of extreme or hazardous hyperbilirubinemia and kernicterus, the etiology of the marked hyperbilirubinemia is often unidentified (1,14,18,19). This unfortunate situation reflects the limited investigative repertoire available to providers in the clinical arena. Once maternal antibody mediated hemolysis and G6PD deficiency are ruled out, the etiology often remains unclear or is conjectured based on the red cell smear and red cell indices. A firm diagnosis is not made. In the US Pilot Kernicterus Registry, idiopathic cases comprised 43% of the total demonstrating a median TSB of 36.0 mg/dL (615 µmol/L) and range of 20.7–52.0 mg/dL (354–889 µmol/L) (14). Few neonates have the capacity to generate TSB levels in a hazardous range; many, if not most have an underlying hemolytic condition (92,93). Christensen and colleagues have utilized a next-generation sequencing gene panel targeted on heritable causes of hemolytic anemia and disorders of hepatic bilirubin uptake and conjugation to clarify the nature of extreme hyperbilirubinemia (92,128) and kernicterus (93). They have recently established a Neonatal Acute Bilirubin Encephalopathy Registry (NABER) that will correlate clinical data with the results of sequencing 28 genes involved in bilirubin production and metabolism to clarify the nature of hyperbilirubinemia in acute bilirubin encephalopathy (129). Molecular diagnosis holds an important key in improving our understanding of the pathogenesis of hazardous hyperbilirubinemia and kernicterus and in identifying means of preventing their occurrence (129).
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (David K. Stevenson and Ronald J Wong) for the series “Neonatal Jaundice” published in Pediatric Medicine. The article has undergone external peer review.
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/pm-21-7). The series “Neonatal Jaundice” was commissioned by the editorial office without any funding or sponsorship. JFW reports serving as a consultant in medico-legal cases related to kernicterus. The author has no other conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bhutani VK, Johnson LH, Maisels MJ, et al. Kernicterus: epidemiologic strategies for its prevention through systems-based approaches. J Perinatol 2004;24:650-62. [Crossref] [PubMed]
- Watchko JF, Lin Z, Clark RH, et al. Complex multifactorial nature of significant hyperbilirubinemia in neonates. Pediatrics 2009;124:e868-77. [Crossref] [PubMed]
- Watchko JF, Lin Z. Genetics of Neonatal Jaundice. In: Stevenson DK, Maisels MJ, Watchko JF, editors. Care of the Jaundiced Neonate. New York: McGraw Hill; 2012:1-27.
- Kaplan M, Hammerman C, Maisels MJ. Bilirubin genetics for the nongeneticist: hereditary defects of neonatal bilirubin conjugation. Pediatrics 2003;111:886-93. [Crossref] [PubMed]
- Kaplan M, Hammerman C. Bilirubin and the genome: the hereditary basis of unconjugated neonatal hyperbilirubinemia. Current Pharmacogenomics 2005;3:21-42. [Crossref]
- Watchko JF, Lin Z. Exploring the genetic architecture of neonatal hyperbilirubinemia. Semin Fetal Neonatal Med 2010;15:169-75. [Crossref] [PubMed]
- Maisels MJ, Gifford K, Antle CE, et al. Jaundice in the healthy newborn infant: a new approach to an old problem. Pediatrics 1988;81:505-11. [PubMed]
- Gale R, Seidman DS, Dollberg S, et al. Epidemiology of neonatal jaundice in the Jerusalem population. J Pediatr Gastroenterol Nutr 1990;10:82-6. [Crossref] [PubMed]
- Maisels MJ, Kring E. Length of stay, jaundice and hospital readmission. Pediatrics 1998;101:995-8. [Crossref] [PubMed]
- Žaja O, Tiljak MK, Stefanovic M, et al. Correlation of UGT1A1 TATA-box polymorphism in breastfed newborns – early presentation of Gilbert’s syndrome. J Matern Fetal Neonatal Med 2014;27:844-50. [Crossref] [PubMed]
- Abdel Fattah M, Ghany EA, Adel A, et al. Glucose-6-phosphate dehydrogenase and red cell pyruvate kinase deficiency in neonatal jaundice cases in Egypt. Pediatr Hematol Oncol 2010;27:262-71. [Crossref] [PubMed]
- Adekunle-Ojo AO, Smitherman HF, Parker R, et al. Managing well-appearing neonates with hyperbilirubinemia in the emergency department observation unit. Pediatr Emerg Care 2010;26:343-48. [Crossref] [PubMed]
- Newman TB, Escobar GJ, Gonzales VM, et al. Frequency of neonatal bilirubin testing and hyperbilirubinemia in a large health maintenance organization. Pediatrics 1999;104:1198-203. [PubMed]
- Johnson L, Bhutani VK, Karp K, et al. Clinical report from the Pilot US Kernicterus Registry (1992 to 2004). J Perinatol 2009;29:S25-S45. [Crossref] [PubMed]
- Manning D, Todd P, Maxwell M, et al. Prospective surveillance study of severe hyperbilirubinemia in the newborn in the UK and Ireland. Arch Dis Child Fetal Neonatal Ed 2007;92:F342-46. [Crossref] [PubMed]
- Sgro M, Campbell D, et al. Incidence and causes of severe neonatal hyperbilirubinemia in Canada. CMAJ 2006;175:587-90. [Crossref] [PubMed]
- Gamaleldin R, Iskander I, Seoud I, et al. An evaluation of risk factors for neurotoxicity in newborns with severe neonatal hyperbilirubinemia. Pediatrics 2011;128:e925-31. [Crossref] [PubMed]
- Alkén J, Håkansson S, Ekéus C, et al. Rates of extreme neonatal hyperbilirubinemia and kernicterus in children and adherence to national guidelines for screening, diagnosis, and treatment in Sweden. JAMA Netw Open 2019;2:e190858. [Crossref] [PubMed]
- Donneborg ML, Hansen BM, Vandborg PK, et al. Extreme neonatal hyperbilirubinemia and kernicterus spectrum disorder in Denmark during the years 2000-2015. J Perinatol 2020;40:194-202. [Crossref] [PubMed]
- Du L, Ma X. International Perspectives: Hyperbilirubinemia and Kernicterus in Neonates in China. NeoReviews 2012;13:e141. [Crossref]
- Crosse VM. The incidence of kernicterus (not due to haemolytic disease) among premature babies. In: Sass-Kortsak A. editor. Kernicterus. Toronto: University of Toronto Press; 1961:4-9.
- Diamond LK, Vaughn VC, Allen FH Jr. Erythroblastosis fetalis. III. Prognosis in relation to clinical and serologic manifestations at birth. Pediatrics 1950;6:630-7. [PubMed]
- Haymaker W, Margoles C, Pentschew A, et al. Pathology of kernicterus and posticteric encephalopathy. In: Kernicterus and its Importance in Cerebral Palsy. A Conference Presented by the American Academy for Cerebral Palsy. Springfield, IL: Charles C Thomas; 1961:21-228.
- Kaplan M, Hammerman C. Glucose-6-phosphate dehydrogenase deficiency and severe neonatal hyperbilirubinemia: a complexity of interactions between genes and environment. Semin Fetal Neonatal Med 2010;15:148-56. [Crossref] [PubMed]
- Preisig D, Bircher J, Preisig R. Positive diagnosis of Gilbert syndrome. Retrospective analysis of 59 cases with special reference to the nicotinic acid test. Schweiz Med Wochenschr 1982;112:1122-9. [PubMed]
- Sieg A, Arab L, Schlierf G, et al. Prevalence of Gilbert’s syndrome in Germany. Dtsch Med Wochenschr 1987;112:1206-8. [Crossref] [PubMed]
- Sun L, Li M, Zhang L, Teng X, et al. Differences in UGT1A1 gene mutations and pathologic liver changes between Chinese patients with Gilbert syndrome and Crigler-Najjar syndrome type II. Medicine (Baltimore) 2017;96:e8620. [Crossref] [PubMed]
- Mi XX, Yan J, Ma XJ, et al. Analysis of the UGT1A1 genotype in hyperbilirubinemia patients: differences in allele frequency and distribution. Biomed Res Int 2019;2019:6272174. [Crossref] [PubMed]
- Johnson L, Garcia ML, Figueroa E, et al. Kernicterus in rats lacking glucuronyl transferase. Am J Dis Child 1961;101:322-49. [Crossref] [PubMed]
- Cannon C, Daood MJ, O’Day TL, et al. Sex-specific regional brain bilirubin content in hyperbilirubinemic Gunn rat pups. Biol Neonate 2006;90:40-5. [Crossref] [PubMed]
- Chaniary KD, Baron MS, Rice AC, et al. Quantification of gait in dystonic Gunn rats. J Neurosci Methods 2009;180:273-7. [PubMed]
- Shaia WT, Shapiro SM, Heller AJ, et al. Immunohistochemical localization of calcium-binding proteins in the brainstem vestibular nuclei of the jaundiced Gunn rat. Hear Res 2002;173:82-90. [Crossref] [PubMed]
- Watchko JF, Daood MJ, Hansen TWR. Brain bilirubin content is increased in P-glycoprotein-deficient transgenic null mutant mice. Pediatr Res 1998;44:763-66. [Crossref] [PubMed]
- Watchko JF, Daood MJ, Mahmood B, et al. P-glycoprotein and bilirubin disposition. J Perinatol 2001;21:S43-S47. [Crossref] [PubMed]
- Bockor L, Bortolussi G, Vodret S, et al. Modulation of bilirubin neurotoxicity by the Abcb1 transporter in the Ugt1-/- lethal mouse model of neonatal hyperbilirubinemia. Hum Mol Genet 2017;26:145-57. [PubMed]
- Daood M, Tsai C, Ahdab-Barmada M, et al. ABC transporter (P-gp/ABCB1, MRP1/ABCC1, BCRP/ABCG2) expression in the developing human CNS. Neuropediatrics 2008;39:211-8. [Crossref] [PubMed]
- Cascorbi I. P-glycoprotein: tissue distribution, substrates, and functional consequences of genetic variations. Handb Exp Pharmacol 2011;201:261-83. [Crossref] [PubMed]
- Hodges LM, Markova SM, Chinn LW, et al. Very important pharmacogene summary: ABCB1 (MDR1, P-glycoprotein). Pharmacogenet Genomics 2011;21:152-61. [Crossref] [PubMed]
- Brites D. The evolving landscape of neurotoxicity by unconjugated bilirubin: role of glial cells and inflammation. Front Pharmacol 2012;3:88. [Crossref] [PubMed]
- Konkle ATM, McCarthy MM. Developmental time course of estradiol. Testosterone, and dihydrotestosterone levels in discrete regions of male and female rat brain. Endocrinology 2011;152:223-35. [Crossref] [PubMed]
- MacLusky NJ, Walters MJ, Clark AS, et al. Aromatase in the cerebral cortex, hippocampus, and mid-brain: ontogeny and developmental implications. Mol Cell Neurosci 1994;5:691-8. [Crossref] [PubMed]
- Dean SL, McCarthy MM. Steroids, sex and the cerebellar cortex: implications for human disease. Cerebellum 2008;7:38-47. [Crossref] [PubMed]
- Bosma PJ, Chowdhury JR, Bakker C, et al. The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 in Gilbert's syndrome. N Engl J Med 1995;333:1171-5. [Crossref] [PubMed]
- Huang MJ, Chen YC, Huang YY, et al. Effect of UDP-glucuronosyltransferase 1A1 activity on risk for developing Gilbert’s syndrome. Kaohsiung J Med Sci 2019;35:432-9. [Crossref] [PubMed]
- Yu WL, Aldrich RA. The glucuronyl transferase system in the newborn infant. Pediatr Clin North Am 1960;7:381-96. [Crossref] [PubMed]
- Valaes T. Bilirubin metabolism: review and discussion of inborn errors. Clin Perinatol 1976;3:177-209. [Crossref] [PubMed]
- Odell GB. Neonatal Hyperbilirubinemia. New York; Grune & Stratton: 1980:41.
- Oski FA. Unconjugated hyperbilirubinemia. In: Avery ME, Taeusch HW. editors. Diseases of the Newborn. Philadelphia: WB Saunders: 1984:630-2.
- Monaghan G, Ryan M, Seddon R, et al. Genetic variation in bilirubin UDP-glucuronosyltransferase gene promoter and Gilbert’s syndrome. Lancet 1996;347:578-81. [Crossref] [PubMed]
- Koiwai O, Nishizawa M, Hasada K, et al. Gilbert’s syndrome is caused by a heterozygous missense mutation in the gene for bilirubin UDP-glucuronosyltransferase. Hum Mol Gen 1995;4:1183-6. [Crossref] [PubMed]
- Aono S, Adachi Y, Uyama E, et al. Analysis of genes for bilirubin UDP-glucuronosyltransferase in Gilbert’s syndrome. Lancet 1995;345:958-9. [Crossref] [PubMed]
- Sugatani J. Function, genetic polymorphism, and transcriptional regulation of human UDP-glucuronosyltransferase (UGT) 1A1. Drug Metab Pharmacokinet 2013;28:83-92. [Crossref] [PubMed]
- Ehmer U, Kalthoff S, Fakundiny B, et al. Gilbert syndrome redefined: a complex genetic haplotype influences the regulation of glucuronidation. Hepatology 2012;55:1912-21. [Crossref] [PubMed]
- Aono S, Yamada Y, Keino H, et al. Identification of defect in the genes for bilirubin UDP-glucuronosyl-transferase in a patient with Crigler-Najjar syndrome type II. Biochem Biophys Res Commun 1993;197:1239-44. [Crossref] [PubMed]
- Sugatani J, Yamakawa K, Yoshinara K, et al. Identification of a defect in the UGT1A1 gene promoter and its association with hyperbilirubinemia. Biochem Biophys Res Commun 2002;292:492-7. [Crossref] [PubMed]
- Yusoff S, Takeuchi A, Ashi C, et al. A polymorphic mutation, c.-3279T>G, in the UGT1A1 promoter is a risk factor for neonatal jaundice in the Malay population. Pediatr Res 2010;67:401-6. [Crossref] [PubMed]
- Beutler E, Gelbert T, Demina A. Racial variability in the UDP-glucuronosyltransferase 1 (UGT1A1) promoter: a balanced polymorphism for regulation of bilirubin metabolism. Proc Natl Acad Sci USA 1998;95:8170-4. [Crossref] [PubMed]
- Sutomo R, Laosombat V, Sadewa AH, et al. Novel missense mutation of the UGT1A1 gene in Thai siblings with Gilbert’s syndrome. Pediatr Int 2002;44:427-32. [Crossref] [PubMed]
- Costa E, Vieira E, Martins M, et al. Analysis of the UDP-glucuronosyltransferase gene in Portuguese patients with a clinical diagnosis of Gilbert and Crigler-Najjar syndromes. Blood Cells Mol Dis 2006;36:91-7. [Crossref] [PubMed]
- Farheen S, Sengupta S, Santra A, et al. Gilbert's syndrome: High frequency of the (TA)7 TAA allele in India and its interaction with a novel CAT insertion in promoter of the gene for bilirubin UDP-glucuronosyltransferase 1 gene. World J Gastroenterol 2006;12:2269-75. [Crossref] [PubMed]
- Sai K, Saeki M, Saito Y, et al. UGT1A1 haplotypes associated with reduced glucuronidation and increased serum bilirubin in irinotecan-administered Japanese patients with cancer. Clin Pharmacol Ther 2004;75:501-15. [Crossref] [PubMed]
- Strassburg CP, Kalthoff S, Ehmer U. Variability and function of family 1 uridi-5’-diphosphate glucuronosyltransferases (UGT1A). Crit Rev Clin Lab Sci 2008;45:485-530. [Crossref] [PubMed]
- Bosma PJ. Inherited disorders of bilirubin metabolism. J Hepatol 2003;38:107-17. [Crossref] [PubMed]
- Long J, Zhang S, Fang X, et al. Association of neonatal hyperbilirubinemia with uridine diphosphate-glucuronosyltransferase 1A1 gene polymorphisms: Meta-analysis. Pediatr Int 2011;53:530-40. [Crossref] [PubMed]
- Yu Z, Zhu K, Wang L, et al. Association of neonatal hyperbilirubinemia with UGT1A1 gene polymorphisms: A meta-analysis. Med Sci Monit 2015;21:3104-14. [Crossref] [PubMed]
- Mehrad-Majd H, Haerian MS, Akhtari J, et al. Effects of Gly71Arg mutation in UGT1A1 gene on neonatal hyperbilirubinemia: a systematic review and meta-analysis. J Matern Fetal Neonatal Med 2019;32:1575-85. [Crossref] [PubMed]
- Bosma PJ, Seppen J, Goldhoorn B, et al. Bilirubin UDP-glucuronosyltransferase 1 is the only relevant bilirubin glucuronidating isoform in man. J Bol Chem 1994;269:17960-64. Erratum: J Biol Chem 1994;269:2542.
- Bancroft JD, Kreamer B, Gourley GR. Gilbert syndrome accelerates development of neonatal jaundice. J Pediatr 1998;132:656-60. [Crossref] [PubMed]
- Hall D, Ybazeta G, Destro-Bisol G, et al. Variability at the uridine diphosphate glucuronosyltransferase 1A1 promoter in human populations and primates. Pharmacogenetics 1999;9:591-9. [Crossref] [PubMed]
- Haverfield EV, McKenzie CA, Forrester T, et al. UGT1A1 variation and gallstone formation in sickle cell disease. Blood 2005;105:968-72. [Crossref] [PubMed]
- Muslu N, Turhan AB, Eskandari G, et al. The frequency of UDP-glucuronosyltransferase 1A1 promoter region (TA)7 polymorphism in newborns and it's relation with jaundice. J Trop Pediatr 2007;53:64-8. [Crossref] [PubMed]
- Kilic I, Cakaloz I, Atalay E. Frequency of UDP-glucuronosyltransferase 1 (UGT1A1) gene promoter polymorphisms in neonates with prolonged and pathological jaundice in the Denizli region of Turkey. Int J Clin Pharmacol Ther 2007;45:475-6. [Crossref] [PubMed]
- Babaoglu MO, Yigit S, Aynacioglu AS, et al. Neonatal jaundice and bilirubin UDP-glucuronosyl transferase 1A1 gene polymorphism in Turkish patients. Basic Clin Pharmacol Toxicol 2006;98:377-80. [Crossref] [PubMed]
- Laforgia N, Faienza MF, Rinaldi A, et al. A. Neonatal hyperbilirubinemia and Gilbert's syndrome. J Perinat Med 2002;30:166-9. [Crossref] [PubMed]
- Carvalho CG, Castro SM, Santin AP, et al. Polymorphic variants of UGT1A1 in neonatal jaundice in southern Brazil. J Trop Pediatr 2010;56:366-7. [Crossref] [PubMed]
- Ulgenalp A, Duman N, Schaefer FV, et al. Analyses of polymorphism for UGT1*1 exon 1 promoter in neonates with pathologic and prolonged jaundice. Biol Neonate 2003;83:258-62. [Crossref] [PubMed]
- Agrawal SK, Kumar P, Rathi R, et al. UGT1A1 gene polymorphisms in North Indian neonates presenting with unconjugated hyperbilirubinemia. Pediatr Res 2009;65:675-80. [Crossref] [PubMed]
- Ergin H, Bican M, Atalay OE. A causal relationship between UDP-glucuronosyltransferase 1A1 promoter polymorphism and idiopathic hyperbilirubinemia in Turkish newborns. Turk J Pediatr 2010;52:28-34. [PubMed]
- Huang MJ, Kua KE, Teng HC, et al. Risk factors for severe hyperbilirubinemia in neonates. Pediatr Res 2004;56:682-9. [Crossref] [PubMed]
- Chou HC, Chen MH, Yang HY, et al. 211 G to A variation of UDP-glucuronosyl transferase 1A1 gene and neonatal breastfeeding jaundice. Pediatr Res 2011;69:170-4. [Crossref] [PubMed]
- Yang H, Wang Q, Zheng L, et al. Multiple genetic modifiers of bilirubin metabolism involvement in significant neonatal hyperbilirubinemia in patients of Chinese descent. PLoS One 2015;10:e0132034. [Crossref] [PubMed]
- Nguyen TT, Zhao W, Yang X, et al. The relationship between hyperbilirubinemia and the promoter region and first exon of UGT1A1 gene polymorphisms in Vietnamese newborns. Pediatr Res 2020;88:940-4. [Crossref] [PubMed]
- Newman TB, Easterling MJ, Goldman ES, et al. Laboratory evaluation of jaundice in newborns. Am J Dis Child 1990;144:364-8. [Crossref] [PubMed]
- Akaba K, Kimura T, Sasaki A, et al. Neonatal hyperbilirubinemia and mutation of the bilirubin uridine diphosphate-glucuronosyltransferase gene: a common missense mutation among Japanese, Koreans and Chinese. Biochem Mol Biol Int 1998;46:21-6. [Crossref] [PubMed]
- Zhou YY, Lee LY, Ng SY, et al. UGTA1A1 haplotype mutation among Asians in Singapore. Neonatology 2009;96:150-5. [Crossref] [PubMed]
- Jirsa M, Petrasek J, Vitek L. Linkage between A(TA)7TAA and -3279T>G mutations in UGT1A1 is not essential for pathogenesis of Gilbert syndrome. Liver Int 2006;26:1302-3. [Crossref] [PubMed]
- Costa E, Viera E, dos Santos R. The polymorphism c.-3279T>G in the phenobarbital responsive enhancer module of the bilirubin UDP-glucuronosyltransferase gene is associated with Gilbert syndrome. Clin Chem 2005;51:2204-6. [Crossref] [PubMed]
- Li Z, Song L, Hao L. The role of UGT1A1 (c.-3279T>G) gene polymorphisms in neonatal hyperbilirubinemia susceptibility. BMC Med Genet 2020;21:218. [Crossref] [PubMed]
- Cheung TP, Rostenberghe HV, Ismail R, et al. High resolution melting analysis of the NR1I3 genetic variants: is there an association with neonatal hyperbilirubinemia? Gene 2015;573:198-204. [Crossref] [PubMed]
- Chen S, Tukey RH. Humanized UGT1 mice, regulation of UGT1A1, and the role of the intestinal tract in neonatal hyperbilirubinemia and breast milk-induced jaundice. Drug Metab Dispos 2018;46:1745-55. [Crossref] [PubMed]
- Maruo Y, Nakahara S, Yanagi T, et al. Genotype of UGT1A1 and phenotype correlation between Crigler-Najjar syndrome type II and Gilbert syndrome. J Gastroenterol Hepatol 2016;31:403-8. [Crossref] [PubMed]
- Christensen RD, Nussenzveig RH, Yaish HM, et al. Causes of hemolysis in neonates with extreme hyperbilirubinemia. J Perinatol 2014;34:616-9. [Crossref] [PubMed]
- Christensen RD, Agarwal AM, George TI, et al. Acute neonatal bilirubin encephalopathy in the State of Utah 2009-2018. Blood Cells Mol Dis 2018;72:10-3. [Crossref] [PubMed]
- Kaplan M, Renbaum P, Levy-Lahad E, et al. Gilbert syndrome and glucose-6-phosphate dehydrogenase deficiency: a dose-dependent genetic interaction crucial to neonatal hyperbilirubinemia. Proc Natl Acad Sci U S A 1997;94:12128-32. [Crossref] [PubMed]
- Huang CS, Chang PF, Huang MJ, et al. Glucose-6-phosphate dehydrogenase deficiency, the UDP-glucuronosyl transferase 1A1 gene, and neonatal hyperbilirubinemia. Gastroenterology 2002;123:127-33. [Crossref] [PubMed]
- Kaplan M, Renbaum P, Vreman HJ, et al. (TA)n UGT 1A1 promoter polymorphism: a crucial factor in the pathophysiology of jaundice in G-6-PD deficient neonates. Pediatr Res 2007;61:727-31. [Crossref] [PubMed]
- Berardi A, Lugli L, Ferrari F, et al. Kernicterus associated with hereditary spherocytosis and UGT1A1 promoter polymorphism. Biol Neonate 2006;90:243-6. [Crossref] [PubMed]
- Iolascon A, Faienza MF, Moretti A, et al. UGT1 promoter polymorphism accounts for increased neonatal appearance of hereditary spherocytosis. Blood 1998;91:1093. [Crossref] [PubMed]
- Kaplan M, Hammerman C, Renbaum P, et al. Gilbert’s syndrome and hyperbilirubinemia in ABO incompatible neonates. Lancet 2000;356:652-3. [Crossref] [PubMed]
- Sampietro M, Lupica L, Perrero L, et al. The expression of uridine diphosphate glucuronosyltransferase gene is a major determinant of bilirubin level in heterozygous beta-thalassaemia and in glucose-6-phosphate dehydrogenase deficiency. Br J Haematol 1997;99:437-9. [Crossref] [PubMed]
- Kang LL, Liu ZL, Zhang HD. Gilbert’s syndrome coexisting with hereditary spherocytosis might not be rare: Six case reports. World J Clin Cases 2020;8:2001-8. [Crossref] [PubMed]
- Maisels MJ, Newman TB. The epidemiology of neonatal hyperbilirubinemia. In: Stevenson DK, Maisels MJ, and Watchko JF, editors. Care of the Jaundiced Neonate. New York: McGraw Hill; 2012:97-113.
- Flaherman VJ, Maisels MJ. Academy of Breastfeeding Medicine. ABM Clinical Protocol #22: Guidelines for Management of Jaundice in the Breastfeeding Infant 35 Weeks or More of Gestation-Revised 2017. Breastfeed Med 201712:250-7.
- Monaghan G, McLellan A, McGeeban A, et al. Gilbert’s syndrome is a contributory factor in prolonged unconjugated hyperbilirubinemia of the newborn. J Pediatr 1999;134:441-6. [Crossref] [PubMed]
- Maruo Y, Nishizawa K, Sato H, et al. Prolonged unconjugated hyperbilirubinemia associated with breast milk and mutations of the bilirubin uridine diphosphate-glucuronosyltransferase gene. Pediatrics 2000;106:E59. [Crossref] [PubMed]
- Maruo Y, Morioka Y, Fujito H, et al. Bilirubin uridine diphosphate-glucuronosyltransferase variation is the genetic basis of breast milk jaundice. J Pediatr 2014;165:36-41.e1. [Crossref] [PubMed]
- Roy-Chowdhury N, Deocharan B, Bejjanki HR, et al. Presence of the genetic marker for Gilbert syndrome is associated with increased level and duration of neonatal jaundice. Acta Paediatr 2002;91:100-1. [Crossref] [PubMed]
- Ramos A, Silverberg M, Stern L. Pregnanediols and neonatal hyperbilirubinemia. Am J Dis Child 1966;111:353-6. [PubMed]
- Fujiwara R, Chen S, Karin M, et al. Reduced expression of UGT1A1 in intestines of humanized UGT1 mice via inactivation of NF-κB leads to hyperbilirubinemia. Gastroenterology 2012;142:109-118.e2. [Crossref] [PubMed]
- Fujiwara R, Maruo Y, Chen S, et al. Role of extrahepatic UDP-glucuronosyl-transferase 1A1: Advances in understanding breast milk-induced neonatal hyperbilirubinemia. Toxicol Appl Pharmacol 2015;289:124-32. [Crossref] [PubMed]
- Strassburg CP, Kneip S, Topp J, et al. Polymorphic gene regulation and interindividual variation of UDP-glucuronosyl-transferase activity in human small intestine. J Biol Chem 2000;275:36164-71. [Crossref] [PubMed]
- Ohno S, Nakajin S. Determination of mRNA expression of human UDP-glucuronosyltransferases and application for localization in various human tissues by real-time reverse transcriptase-polymerase chain reaction. Drug Metab Dispos 2009;37:32-40. [Crossref] [PubMed]
- Alonso EM, Whitington PF, Whitington SH, et al. Enterohepatic circulation of nonconjugated bilirubin in rats fed with human milk. J Pediatr 1991;118:425-30. [Crossref] [PubMed]
- Gartner LM. Breastfeeding and jaundice. J Perinatol 2001;21:S25-S29. [Crossref] [PubMed]
- Fevery J, Blanckaert N, Heirwegh KP, et al. Unconjugated bilirubin and an increased proportion of bilirubin monoconjugates in the bile of patients with Gilbert’s syndrome and Crigler-Najjar disease. J Clin Invest 1977;60:970-9. [Crossref] [PubMed]
- Muraca M, Fevery J, Blanckaert N. Relationship between serum bilirubins and production and conjugation of bilirubin. Studies in Gilbert’s syndrome, Crigler-Najjar disease, hemolytic disorders and rat models. Gastroenterology 1987;92:309-17. [Crossref] [PubMed]
- Spivak W, DiVenuto D, Yuey W. Non-enzymatic hydrolysis of bilirubin mono- and diglucuronide to unconjugated bilirubin in model and native bilirubin systems. Biochem J 1987;242:323-9. [Crossref] [PubMed]
- Hsieh TY, Shiu TY, Huang SM, et al. Molecular pathogenesis of Gilbert’s syndrome: decreased TATA-binding protein binding affinity of UGT1A1 gene promoter. Pharmacogenet Genomics 2007;17:229-36. [Crossref] [PubMed]
- Premawardhena A, Fisher CA, Liu YT, et al. The global distribution of length polymorphisms of the promoters of the glucuronosyltransferase 1 gene (UGT1A1): hematologic and evolutionary implications. Blood Cells Mol Dis 2003;31:98-101. [Crossref] [PubMed]
- Seppen J, Bosma P. Bilirubin, the gold within. Circulation 2012;126:2547-9. [Crossref] [PubMed]
- Wasser DE, Hershkovitz I. The question of ethnic variability and the Darwinian significance of physiologic neonatal jaundice in East Asian populations. Med Hypotheses 2010;75:187-9. [Crossref] [PubMed]
- Gazzin S, Vitek L, Watchko JF, et al. A novel perspective on the biology of bilirubin in health and disease. Trends Mol Med 2016;22:758-68. [Crossref] [PubMed]
- Lin Z, Fontaine J, Watchko JF. Co-expression of gene polymorphisms involved in bilirubin production and metabolism. Pediatrics 2008;122:e156-e162. [Crossref] [PubMed]
- D’Silva S, Colah RB, Ghosh K, et al. Combined effects of the UGT1A1 and OATP2 gene polymorphisms as major risk factor for unconjugated hyperbilirubinemia in Indian neonates. Gene 2014;547:18-22. [Crossref] [PubMed]
- Huang MJ, Lin YC, Liu K, et al. Effects of variation status and enzyme activity for UDP-glucuronosyltransferase A1A gene on neonatal hyperbilirubinemia. Pediatr Neonatol 2020;61:506-12. [Crossref] [PubMed]
- Vockley J, Rinaldo P, Bennett MJ, et al. Synergistic heterozygosity: disease resulting from multiple partial defects in one or more metabolic pathways. Mol Genet Metab 2000;71:10-8. [Crossref] [PubMed]
- Zangen S, Kidron D, Gelbart T, et al. Fatal kernicterus in a girl deficient in glucose-6-phosphate dehydrogenase: a paradigm of synergistic heterozygosity. J Pediatr 2009;154:616-9. [Crossref] [PubMed]
- Rets A, Clayton AL, Christensen RD, et al. Molecular diagnostic update in hereditary hemolytic anemia and neonatal hyperbilirubinemia. Int J Lab Hematol 2019;41:95-101. [PubMed]
- Bahr TM, Christensen RD, Agarwal AM, et al. The Neonatal Acute Bilirubin Encephalopathy Registry (NABER): Background, aims, and protocol. Neonatology 2019;115:242-6. [Crossref] [PubMed]

